- Open access
- Published: 13 August 2024

Microbial remediation of polluted environment by using recombinant E. coli : a review
- Samriti Sharma 1 ,
- Shruti Pathania 2 ,
- Suhani Bhagta 3 ,
- Neha Kaushal 2 ,
- Shivani Bhardwaj 4 ,
- Ravi Kant Bhatia 5 &
- Abhishek Walia 4
Biotechnology for the Environment volume 1 , Article number: 8 ( 2024 ) Cite this article
420 Accesses
Metrics details
An increased amount of toxins has collected in the environment (air, water, and soil), and traditional methods for managing these pollutants have failed miserably. Advancement in modern remediation techniques could be one option to improve bioremediation and waste removal from the environment. The increased pollution in the environment prompted the development of genetically modified microorganisms (GEMs) for pollution abatement via bioremediation. The current microbial technique focuses on achieving successful bioremediation with engineered microorganisms. In the present study, recombination in E. coli will be introduced by either insertion or deletion to enhance the bioremediation properties of the microbe. Bioremediation of domestic and industrial waste performed using recombinant microbes is expensive but effectively removes all the waste from the environment. When compared to other physicochemical approaches, using microbial metabolic ability to degrade or remove environmental toxins is a cost-effective and safe option. These synthetic microorganisms are more effective than natural strains, having stronger degradative capacities and the ability to quickly adapt to varied contaminants as substrates or co-metabolites. This review highlights the recent developments in the use of recombinant E. coli in the biodegradation of a highly contaminated environment with synthetic chemicals, petroleum hydrocarbons, heavy metals, etc. It also highlights the mechanism of bioremediation in different pollution sources and the way in which this genetically altered microbe carries out its function. Additionally, addressed the benefits and drawbacks of genetically engineered microbes.
Introduction
The advent of global industrialization has brought about critical environmental challenges with pollution being a significant concern. While industrialization has contributed significantly to economic development, technological advancements, and improved living standards for human being, but simultaneously, it has also led to adverse environmental impacts particularly in the context of pollution of the environment [ 1 ]. Environmental pollution refers to the degradation of the natural environment because of the introduction of pollutants. There are several types of environmental pollution including air pollution, water pollution, and soil pollution [ 2 , 3 , 4 , 5 ]. Pollutants encompass substances that, while sometimes naturally occurring, are deemed contaminants when surpassing natural levels. Pollutants can be categorized into biodegradable and nonbiodegradable types. Biodegradable pollutants, like phosphates and organic waste, can be broken down by living organisms. In contrast, nonbiodegradable pollutants, such as plastics, metals, pesticides, glass, and radioactive isotopes, resist decomposition by living organisms, persisting in the ecosphere for extended periods [ 6 ].
Environmental pollution, a ubiquitous and pressing issue, casts a looming shadow over the planet, threatening the delicate balance of ecosystems and endangering the health of both flora and fauna, including humans. From air and water pollution to soil contamination, the consequences of human activities on the environment are manifold and far-reaching [ 7 ]. The call for India to prioritize environmental protection amid its rich biodiversity and stark socio-economic disparities has never been more urgent. Joutey et al. and Rabani et al. [ 8 , 9 ] underscore the critical need for India to balance economic development with environmental conservation. The government of India has taken major steps to prevent pollution in our country (Table 1 ). Addressing environmental pollution requires a combination of regulations, technological advancements, public awareness, and sustainable practices to minimize and mitigate the impact of pollutants on the planet. Numerous laws have been enacted to tackle the escalating pollution levels and set emission standards [ 10 ]. These legislative measures represent crucial milestones in India’s environmental stewardship journey.
Dealing with pollution is a complex challenge, and various methods, both physical and chemical, have been employed to address the pervasive environmental issues (Table 2 ). However, their effectiveness and cost often limit their widespread use. Natural solutions, while safe and effective, face challenges due to the rapid accumulation of pollution from industrialization and the presence of nonbiodegradable synthetic materials [ 11 , 12 ]. Physical methods, such as filtration and soil excavation, can be time-consuming and costly. Chemical alternatives, on the other hand, may pose inherent dangers to the environment and human health. Moving forward, it is imperative for India to adopt a holistic approach to environmental conservation—one that integrates environmental considerations into all aspects of policymaking and development planning. This approach should prioritize the protection of ecosystems, biodiversity, and public health while fostering sustainable economic growth and social equity. Advances in science and technology play a crucial role in pollution mitigation.
In response to these limitations, bioremediation has emerged as a promising and environmentally friendly approach. Bioremediation involves the use of microorganisms to assimilate, digest, or transform hazardous substances into less harmful or nontoxic forms [ 13 , 14 , 15 ]. Microorganisms exhibit remarkable capabilities in degrading, detoxifying, and even accumulating toxic organic and inorganic substances [ 16 , 17 ]. The use of genetically modified organisms (GMOs), such as the genetically modified Escherichia coli , has become a powerful tool in the field of bioremediation. These engineered microorganisms are designed to efficiently remove toxins that indigenous bacteria may struggle to break down, offering a targeted and effective approach to environmental cleanup [ 18 ]. In contemporary bioremediation methods, genetically modified organisms play a pivotal role in addressing environmental pollution, particularly in situations where natural bacterial populations are insufficient to handle specific pollutants. The introduction of a foreign gene into bacteria transforms them into unique strains with enhanced capabilities for rapidly breaking down pollutants, such as hydrocarbons, in the environment [ 19 ]. The use of genetically modified E. coli in bioremediation has several advantages such as precision, efficiency, and versatility. However, it is essential to consider potential ethical and ecological concerns associated with the release of genetically modified organisms into the environment. Robust containment measures and thorough risk assessments are crucial to prevent unintended consequences. Therefore, in this article, the use of genetically modified E. coli in bioremediation is discussed which exemplifies the intersection of biotechnology and environmental science, offering innovative solutions to address pollution challenges. As technology continues to advance, the application of genetic engineering in bioremediation holds significant promise for developing tailored and efficient approaches to environmental cleanup.
- Bioremediation
To deal with pollution, a variety of methods (physical and chemical) are available. Due to their high cost and low effectiveness, most of them are of limited use. Physical methods are time-consuming and expensive, whereas chemical alternatives are inherently dangerous. Natural solutions are safe and effective, although they are sluggish and becoming less effective as a result of industrialization’s rapid pollution buildup and nonbiodegradable synthetic materials [ 11 , 12 ]. Bioremediation is gradually becoming the standard method for restoring contaminated with heavy metals because it is efficient and cost-effective technology for the transformation of contaminants [ 13 , 14 , 15 ]. Biodegradation is a series of chemical reactions that occur in the presence of living organisms such as bacteria, fungi, yeast, algae, and insects in an environment with optimal light, temperature, and oxygen [ 20 ]. Microbes mitigate heavy metals and improve soil fertility, and plant development makes them more preferable source for bioremediation. The molecular nature, gene and enzyme induction, metabolite production, growth efficiency, and survival rate all influence individual bacteria’ potential to act as bioremediation agents [ 21 ]. At higher moisture rate, anaerobic condition persists which slow down the degradation rate. In cold condition, microbial degradation of heavy metal is slow, as metabolic activities are inhibited as the microbial transport routes are frozen by the sub-zero water [ 22 , 23 ]. Similarly, at higher temperature, the rate of heavy metal solubility increases, which increases their availability and the rate of microbial biodegradation [ 24 ]. The rate of microbial biodegradation is determined by the metal or pollutant’s chemical structure, bioavailability, concentration, toxicity, and stability. The degradation of the n-alkanes is more effortless in comparison to the branched alkanes, aromatics with low molecular weight, hydrocarbons with high molecular weight, and the asphaltenes [ 25 ]. Molecular mechanisms play a crucial role in deciphering the microbial metabolism, genes, characteristics, variety, and fluctuations of microorganisms engaged in microbial remediation. Metabolic and protein analysis, sequencing, and the utilization of sophisticated bioinformatics tools are specifically employed to decipher the various categories of microorganisms and the factors influencing them in the bioremediation process [ 23 ].
Microorganisms currently employed in bioremediation have the potential to be genetically engineered in order to augment their enzymatic production, thereby amplifying their capacity for biodegradation. These organisms’ genetic architecture makes them useful for biodegradation, biotransformation, biosorption, and bioaccumulation [ 26 ] (Fig. 1 ). The use of recombinant DNA allows an organism to develop the ability to digest a xenobiotic via degradative genes. Recombinant microorganisms and genetically modified microbes have been used as an effective technique for pollution breakdown [ 27 ]. In the current bioremediation technique, genetically modified organisms are employed to efficiently eliminate pollutants that native bacteria are unable to decompose [ 18 ]. There are varieties of bacteria reported to be capable of feeding on hydrocarbons under anaerobic and aerobic conditions [ 28 ]. Toxic substances may be converted to nontoxic ones by the bioremediation process by a variety of bacteria species such as Achromobacter , Pseudomonas , Dehalococcoides , Rhodococcus , Comamonas , Burkholderia , Alcaligenes , Bacillus subtilis , Aspergillus niger , Deinococcus radioduran , Acidithiobacillus ferrooxidans , Mesorhizobium huakuii , Pseudomonas K-62, Ralstonia , Rhodopseudomonas palustris , and Sphingomonas [ 29 ]. Similarly, nitrate-reducing bacterial strains, Brevibacillus sp. and Pseudomonas sp., were identified in petroleum-contaminated soil. Bacillus , Corynebacterium , Staphylococcus , Streptococcus , Shigella , Alcaligenes , Acinetobacter , Escherichia , Klebsiella , and Enterobacter were the best hydrocarbon-degrading bacteria [ 30 ].

Different approaches adapted by the microbes for the degradation of toxic compounds
Microbe’s genetic sequences have been manipulated keeping specific goal in mind [ 31 ]. The term “genetically engineered organisms” (GEMs) refers to microorganisms (bacteria, fungi, and yeast, among others) that have been altered by humans utilizing molecular biology in vitro procedures [ 32 ]. There has been an explosion in the expansion of genetic engineering and recombinant DNA in breeding microorganisms, resulting in a huge number of bacteria with effective engineering that boosted pollutant-degrading abilities [ 18 , 27 , 33 ]. Bioremediation research is awaiting the introduction of gene editing technologies that produce knock-in and knockout. According to recent articles, researchers have mostly used the CRISPR-Cas system with model organisms such as Pseudomonas or E. coli [ 34 ]. Even non-model organisms like Achromobacter sp. HZ01 and Comamonas testosteroni may be employed for bioremediation due to new insights into CRISPR tools and the synthesis of gRNA to express function-specific genes pertinent to remediation [ 35 , 36 ]. In experiments involving organophosphate and pyrethroid bioremediation, genetically altered Pseudomonas putida KT2440 was employed [ 37 ]. White rot fungus produces enzymes that break down polycyclic aromatic hydrocarbons (PAHs), TNT (2,4,6-trinitrotoluene), and polycyclic aromatic hydrocarbons (PCBs). When the enzyme esterase D combines with the insecticide endosulfan (an organochlorine), it produces simpler molecules. LiP-encoded hemoproteins in Phanerochaete chrysosporium degrade PAHs [ 38 ].
Recombinant E. coli in bioremediation
E. coli is a rod-shaped facultative coliform bacterium belonging to the genus Escherichia that measures only about 1 µm long by 0.35 µm wide. It is one of the model organisms used in bioremediation (Fig. 2 ). E. coli is generally known as the “work horse” of molecular biology for its fast growth rate in chemically defined media and the various tools available for its genetic modifications. E. coli harbors a genome with features like an organized structure, a remnant of many phages, insertion sequences (IS), and high transport capacity towards the cytoplasm [ 39 ]. E. coli is a preferred host for gene cloning due to the ease with which DNA molecules may be introduced into the cells. Protein production in E. coli is expected due to the strain’s rapid growth and high protein expression levels [ 40 ]. Various studies show that enteric bacterium like E. coli form phenol and p-cresol when grown on natural media (peptone and casein media) as well as in chemically defined media, i.e., L-tyrosine and p-hydroxybenzoic acid media [ 41 ]. According to a study conducted by Burlingame and Chapman [ 42 ] in 1983, it was found that E. coli has the capability to mineralize several aromatic acids, such as PA (phenylacetic acid), HPA (hydroxyphenylacetic acid), PP (phenyl propionic acid), 3HPP (hydroxyl phenyl propionic acid), and 3HCl. These research findings emphasized the ability of E. coli to break down and utilize a diverse range of aromatic acids [ 22 ]. An E. coli bacterium that has been genetically modified is employed as a highly effective agent in the process of bioremediation. The incorporation of a gene into bacteria results in the conversion of the bacteria into a distinct strain that possesses the ability to efficiently eliminate hydrocarbon pollutants from the surrounding environment (Fig. 3 ) [ 19 ]. There exist multiple methods for manipulating microbial genetics through genome editing, each of which is quite efficient and has been used in E. coli genome editing, making it capable of degrading pollutants and converting them to less harmful molecules [ 43 ]. The curli of an E. coli cell was genetically modified to produce BIND-PETase [ 40 ]. The E. coli SE5000 strain underwent genetic modification by introducing the nixA gene, which enables the expression of a nickel transporting system. This system has the ability to degrade nickel from aqueous system [ 44 , 45 ]. The E. coli FACU strain possesses a significant capacity to reduce Cr (IV) to Cr (III) exhibiting great potential as a viable agent in the bioremediation of hazardous chromium species in aerobic environmental conditions [ 46 ].

Mechanism of biosorption on the basis of cell metabolism and its location within cell or metal removable

Overview of bioremediation methods
General mechanism of degradation of pollutant by recombinant microbe
Genetic manipulation possesses the ability to create or mend microorganisms, leading to the development of biological detection systems that exhibit enhanced internal robustness, specificity, and resilience in various environments. Genetically engineered microorganisms (GEM) refer to microorganisms that have undergone genetic modifications using techniques of genetic engineering (inspired by the natural genetic exchange observed between microorganisms) [ 47 , 48 ]. GEMs (genetically engineered microbes) have shown promise in the bioremediation of soil, groundwater, and activated sludge, with improved degrading capabilities for a variety of chemical contaminants [ 28 ]. Microbes possess inherent biological mechanisms that enable them to withstand intense metal stress or eradicate metals from their surroundings. Microbial bioremediation employs the following mechanisms [ 49 ]:
(1) Cell wall components or intracellular metal-binding proteins and peptides, such as metallothioneins (MT) and phytochelatins, play a crucial role in sequestering toxic metals. Additionally, substances like bacterial siderophores, which are mainly catecholates, are also involved in this process. It is worth noting that fungi produce hydroxamate siderophores [ 50 ].
(2) Altering metabolic processes directly blocks metal uptake.
(3) Enzymes are used to convert metals into harmless forms.
(4) Efflux mechanisms have the potential to decrease metal levels within the intracellular milieu.
Environmental contaminants such as chlorobenzene acids, toluene, and other halogenated insecticides and toxic wastes are broken down into less harmful forms by using important genes. A different plasmid is required for each chemical [ 51 , 52 ]. Plasmids are classified into four groups [ 53 ].
1) OCT plasmid (degrades octane, hexane, and decane).
2) XYL plasmid (degrades xylene and toluenes).
3) CAM plasmid (degrades camphor).
4) NAH plasmid (degrades naphthalene).
The appearance and dissemination of genes that break down pesticides can yield a beneficial impact on the elimination of hazardous waste from the surroundings. The potency of E. coli in the degradation of various pollutants has been shown in Table 3 . The genetically engineered strain of E. coli is able to express the Hg 2+ and metallothionein transport systems. Excessive exposure to Saccharomyces cerevisiae glutathione S-transferase fusion protein and pea metallothionein significantly increased Hg 2+ expression delivered by MerP and MerT, which protect cells from Hg 2+ [ 54 , 55 ]. Similarly, horizontal gene transfer (HGT) methods have been employed for incorporating petrol-contaminated organisms with E. coli carrying the vector pSF-OXB15-p450 cam fusion, which showed that E. coli bacteria are useful for the degradation of heavy metals [ 56 ]. Recombinant E. coli that expresses the metallothionein gene ( Neurospora crassa ) for Cd uptake was created using plasmid-encoded biochemical information and genetic engineering techniques, yielding a significantly faster Cd uptake than the donor microbe [ 57 ].
Different types of pollution and their bioremediation using recombinant E. coli
Soil contamination.
Soil is an essential ecosystem consisting of both living and nonliving elements. The entirety of the natural world relies on soil in various ways. It serves as a connection between the biosphere, atmosphere, and hydrosphere, thereby playing a crucial role in maintaining the ecological equilibrium [ 67 ]. There exists a disagreement in the definition of “soil contamination.” According to certain viewpoints, soil is deemed contaminated when the concentration of chemicals exceeds its typical range. While some individuals express concerns regarding the establishment of the standard range for pollutants. Hence, it can be asserted that “soil which is unsuitable for utilization and incapable of fulfilling its purpose is deemed as contaminated” [ 68 ]. The quality of soil and its role in ecological balance are affected by the addition of soil contaminants due to natural processes and human activities like industrial wastes, the use of fertilizers in agricultural activities, and domestic and commercial construction [ 69 ]. Broadly, two types of contaminants contribute to soil pollution: inorganic and organic. In the category of inorganic pollutants, heavy metals are placed at the top of the list and are present in most of the contaminated sites. The most common toxic heavy metal contaminants found in soil include mercury (Hg), arsenic (As), copper (Cu), cadmium (Cd), chromium (Cr), Zinc (Zn), lead (Pb), and nickel (Ni) [ 70 ]. These soil contaminants sink into the soil through bad agricultural practices, inefficient industrial effluent disposal techniques, unauthorized waste dumping, etc. The persistence of heavy metals in natural environments presents a more formidable obstacle in comparison to organic contaminants, as they exhibit resistance to both microbial and chemical degradation. As a result, the elimination of heavy metals becomes a long-lasting challenge once they are introduced [ 71 ]. Organic contaminants encompass carbon-containing substances, regardless of the presence or absence of functional groups within their structures. The list of organic contaminants that contribute to soil pollution includes insecticides (e.g., captan, benomyl, endosulfan, heptachlor), herbicides (atrazine, alachlor, acetochlor, etc.), oil hydrocarbons (e.g., alkanes, alkenes), chlorinated compounds (e.g., polychlorinated biphenyls (PCB), polychlorinated dibenzodioxins (PCDD), polychlorinated dibenzofurans (PCDF)), aromatic hydrocarbons (e.g., BTEX, i.e., benzene, toluene, ethylbenzene, xylene), biocides (benzalkonium chloride), and polycyclic dibenzo-p-dioxins (e.g., benzopyrene, chrysene, fluoranthene) [ 72 ]. Persistent organic pollutants (POPs) are classified as organic contaminants, which are regarded as the most prioritized category of organic contaminants due to their high toxicity, carcinogenic properties, and ability to bioaccumulate in the environment [ 73 ]. Taking this into consideration, numerous nations have implemented limitations or outright prohibited the utilization and production of persistent organic pollutants (POPs). The POP compounds encompass substances such as DDT, endrin, hexachlorobenzene, PCBs, PCDD, PCDF, and others [ 68 , 74 ] (Fig. 4 ).

Direct enzymatic and indirect mobilization of radionuclides by metal-reducing microorganisms via capturing of electrons derived by organic compounds (lactate and acetate)
Traditionally, various techniques are used to remove the soil contaminants, including extraction and separation techniques, thermal methods, chemical methods, microbial treatment methods, solid waste treatments, and phytoremediation (Table 4 ). The existing treatments for soil pollution, as discussed above, are not very successful in the removal of contaminants; sometimes, they bring down the concentration of contaminants at the cost of soil quality. Some of the techniques are also less cost-effective [ 75 , 76 ]. The main objective of soil remediation is not just the elimination of contaminants but also to restore the quality of the soil. So, we need to shift towards a new approach that gives better and more desirable results in terms of the elimination of pollutants and the restoration of soil quality [ 77 ]. Bioremediation is one of those approaches on which we can rely. In recent years, bioremediation has emerged as a great alternative to existing treatments as it is economical and does not compromise the health of the soil [ 78 ].
Recombinant E. coli strain used for bioremediation of soil
The various studies done by Almaguer-Cantú et al. [ 80 ] on the removal of heavy metal contaminants from soil reported that genetically modified E. coli cells with overexpression of pea metallothionein MT improve the biosorption of Ni 2+ and efficiently remove Ni 2+ contamination from the affected sites. The elimination of hazardous metals from a polluted area through the utilization of biosorbent cell surface components of microorganisms is known as biosorption [ 81 ]. These biosorbent cell surface moieties are present on the outer surfaces of fungi, algae, and bacteria. Bacteria are widely regarded as the superior biosorbent when compared to other microorganisms [ 82 ]. This is primarily attributed to their possession of chemosorption sites such as teichoic acid, as well as their remarkable surface-to-volume ratio. These characteristics greatly enhance their biosorption capabilities [ 83 ]. In a study done in the United States for the removal of atrazine contamination from the contaminated fields by using recombinant E. coli encapsulating AtzA, which is responsible for the degradation of atrazine, they observed that after 8 weeks of inoculation, atrazine levels decreased by 52% and 77% (Table 5 ) in plots containing killed recombinant E. coli cells and combinations of phosphate, respectively [ 63 , 84 ].
The successful elimination of oil contaminants in soil caused by oil spills can be achieved through the introduction of genetically modified E. coli cells containing catabolic genes [ 92 ]. The overexpression of three enzymes, namely almA, xylE, and p450cam, results in the degradation of petroleum hydrocarbon. According to their research, this genetically modified E. coli was able to decrease the level of petroleum hydrocarbon concentration by as much as 46% after a period of 60 days following inoculation [ 56 ]. Mercury (Hg) is a hazardous heavy metal and a significant inorganic pollutant found in soil, which can have harmful consequences on the organisms inhabiting contaminated areas. When it infiltrates the human body through the food chain, it gives rise to serious ailments such as neural disorders and respiratory disorders, occasionally leading to fatality. Genetically modified E. coli JM109 cells can assist in eliminating the Hg 2+ contamination present in the soil [ 93 ]. This strain of E. coli has been genetically modified to produce the merT-merP protein and metallothionein, which are responsible for the accumulation of Hg 2+ in the organism [ 64 ]. E. coli SE5000, a genetically modified strain, possesses the GSM-MT and nixA genes. The nixA gene is accountable for the activation of the Ni 2+ transport system, enabling it to effectively eliminate Ni 2+ contamination. On the other hand, GSM-MT is responsible for the increased production of metallothionein in the form of a glutathione S-transferase fusion protein [ 94 ].
Air pollution
Despite the remarkable advancements in technology, society, and the provision of various services, the Industrial Revolution had a detrimental impact on human health due to the significant release of pollutants into the air ( http://www.who.int/airpollution/en/ ). Air pollution is the term used to describe the existence of detrimental substances in the atmosphere of our planet, which has adverse effects on both human well-being and the environment [ 95 , 96 ]. The increase in economic growth has been accomplished by elevated energy consumption. The rapid urbanization in India, coupled with swift economic progress, has led to a surge in air pollution levels within megacities [ 97 ]. Particulates, greenhouse gases, and smog-forming substances such as sulfur dioxide (SO 2 ), ground-level ozone (O 3 ), nitrogen oxides (NO 2 ), and volatile organic compounds, are all major air pollutants (VOCs) [ 98 , 99 ]. Air pollution has adverse effects not only on humans but also on the marine environment and is responsible for climate change too. The degradation of the earth’s atmosphere is closely linked to the relationship between climate change and air pollution. The elevated concentrations of methane, black carbon, aerosols, and tropospheric ozone disturb the incoming solar radiation. Consequently, the temperature is on the rise, leading to the melting of icebergs, ice, and glaciers [ 22 , 100 ]. The World Health Organization provides information on different categories of air pollutants, such as particle pollution, ground-level ozone, carbon monoxide, sulfur oxides, nitrogen oxides, and lead. In 2011, Delhi recorded a PM10 level of 198 μg m −3 , which exceeds the minimum limit by a factor of 10 [ 101 , 102 ]. In May 2014, the city of New Delhi earned the unfortunate distinction of being the most polluted city in the world, according to the World Health Organization (WHO). This was primarily attributed to the high concentration of particle matter (PM) with a diameter less than 2.5 µm, which exceeded 350 µg per cubic meter of air in New Delhi. ( http://www.theguardian.com/news/datablog/2015/jun/24/air-pollution-delhi-is-dirty-but-how-do-other-cities-fare ) [ 95 ]. A conference titled “Impact of Disease: Air Pollution as a Leading Cause” was organized in New Delhi on February 13, 2013, by the Centre for Science and Environment (CSE) in collaboration with the Health Effects Institute, Boston, USA, and the Indian Council of Medical Research, New Delhi ( http://www.cseindia.org/content/workshop-global-burden-disease-air-pollution-amongst-top-killers-india ).
Air pollution can be easily dispersed and transported between different areas. This detrimental pollution leads to significant issues for both the environment and human health. Consequently, it is imperative to discover effective decontamination strategies in order to cleanse the environment. The process of decontamination must be carried out in a manner that safeguards the well-being of both animals and humans while also promoting the circulation of clean air [ 31 ] (Perera and Hemamali, 2022). Consequently, there is an increasing desire to discover efficient methods for remediating polluted areas, whether partially or entirely, in order to restore their environmental integrity [ 103 , 104 ].
Degradation of air pollutant by recombinant E. coli microbe
Bacteria facilitate the breakdown of dangerous chemicals through an assimilative mechanism, wherein they acquire carbon and energy to support their growth, ultimately leading to the conversion of the compound into minerals [ 105 , 106 ]. The bacteria responsible for PAH degradation include Achromobacter sp . , Bacillus sp., Mycobacterium sp . , Burkholderia sp., Pseudomonas sp., Rhodococcus sp., Stenotrophomonas maltophilia , Sphingomonas sp., Xanthomonas sp., and Xanthomonas sp. [ 107 , 108 ]. The initial stage of hydrocarbon degradation involves the transformation of polycyclic aromatic hydrocarbon (PAH) or alkane chain into basic alcohol, subsequently converting into aldehyde and ultimately resulting in water, carbon dioxide, and biomass through oxidation. Oxidation also leads to the conversion of reduced sulfur molecules like H 2 S into inorganic sulfur and thiosulfate, forming corrosive sulfuric compounds [ 101 , 109 ]. H 2 S advance oxidation is completed by chemolithotrophs. Sulfate is ingested through the sulfate start pathway, which is made up of three responses: adenosine 5′-phosphorylation of APS, GTP hydrolysis, and APS 3′-phosphorylation to deliver 3′-phosphoadenosine 5′-phosphosulfate (PAPS) [ 110 , 111 ]. Microbes that degrade sulfate have the ability to use hydrocarbons and hydrolyze complicated chemicals in soil, according to Rennenberg [ 112 ]. Besides, a designed strain able of debasing PAHs was made in E. coli by communicating salicylate oxygenase, a protein encoded by bphA2cA1c from Sphingomonas yanoikuyae B1 [ 113 ]. The pGEX-AZR/ E. coli JM-109 strain was genetically engineered, resulting in enhanced efficiency for decomposing various azo dyes [ 114 ].
Water pollution
The express “water contamination” is characterized in an assortment of ways by different committees, with the objective of making strides the quality of our environment. Agreeing to the head of the science committee, Washington, USA, in 1965, characterized water contamination as an alteration within the physical, compound, and organic qualities of water that will cause risky impacts on human and maritime life. Nowadays, it is not only concerned with public health but also with destroying natural beauty, resources, aesthetics, and the conservation of water [ 115 , 116 ]. Numerous anthropogenic exercises are related to water contamination and have driven to water quality disintegration, like industrialization, chemical-related cultivating, broad urbanization, and populace development [ 117 ]. There are two sorts of sources that are included in water contamination, i.e., point sources and nonpoint sources. The coordinate identifiable source, or where coordinate association is appeared, is known as a point source, such as mechanical effluents, oil spills, and metropolitan and mechanical squander water effluents. In terms of nonpoint sources, diverse sources are included within the event of water contamination, primarily urban squander, runoff from rural areas, radioactive water (from atomic reprocessing plants), and contaminants that enter ground-level water [ 49 ]. Water contamination is caused by a variety of factors, the most prominent of which being urbanization (higher phosphorus concentrations in urban catchments sewage waste (massive increase in the growth of algae or plankton that facilitate huge areas of oceans, lakes, or rivers), industrial waste (wastes containing acids, alkalis, dyes, and other chemicals), agro-chemical waste (include fertilizers, pesticides which may be herbicides and insecticides), nutrient enrichment, thermal pollution (nuclear power and electric power plants, petroleum refineries, steel melting factories, coal fire power plant, boiler from industries), oil spillage (petrol, diesel, and their derivatives pollute seawater), acid rain pollution, and radioactive pollution (radioactive sediment, waters used in nuclear atomic plants, radioactive minerals exploitation, nuclear power plants) [ 65 , 118 , 119 ].
Water contamination is treated using a variety of physical and chemical approaches [ 75 , 94 ]. Screening (radioactive sediment, waters used in nuclear atomic plants, radioactive mineral exploitation, nuclear power plants), grit chamber (remove sand and egg shells), floatation (oils, fats, grease, sediment solids), and sedimentation tank clarifier are examples of physical treatments (remove heavier sludge solids), whereas chemical treatments are as follows: neutralization (it adjusts pH for maintaining acidity of water), flocculation, coagulation (solid removal, water clarification, lime softening by chemical flocculants and coagulants), oxidation (may reduce toxicity using biochemical oxygen demand), ozonation (degradation of organic and inorganic pollutants), and chlorination [ 115 , 120 ].
Drawbacks of physical and chemical methods
So many by-products are formed, chemical consumption is so high, physicochemical monitoring of effluents, capital and energy costs are so high, high sludge production and management of disposable, and techniques are expensive and toxic to the environment (Table 6 ).
Biological method used for water treatment
The biological method is the most common sanitizing method used for wastewater treatment, and it is also called secondary treatment, which involves the removal of organic matter from wastewater using bacteria and other microorganisms [ 137 ]. Wastewater typically contains pathogenic organisms, heavy metals, toxins, and organic matter (garbage, waste, and partially digested foods) [ 138 ]. Biological methods can be classified into two categories: (i) aerobic—takes place in the presence of oxygen and (ii) anaerobic—takes place in the absence of oxygen. Aerobic biological treatment involves many processes, i.e., the activated sludge process, trickling filters, aerated lagoons, and oxidation ponds. Due to its ease of use, rapidity, and efficiency, this process removes up to 98% of organic contaminants. Anaerobic biological treatment is used to treat high-strength wastewater (sludge degradation and stabilization). The process is slow as compared to aerobic; biogas production is one example of biodegradation of material where it overall converts up to 60% of organic solid mass ( http://neoakruthi.com/blog/biological-treatment-of-wastewater.html ) [ 139 ].
Some of the examples of microorganisms that are involved in the treatment of wastewater using different processes are gram-negative bacteria (proteobacteria) for the elimination of organic elements and nutrients, Bacillus , Bacteroidetes, Acidobacteria, Chloroflexi, Tetrasphaera , Trichococcus , Rhodobacter , Pseudomonas , E. coli , Hyphomicrobium ascomycetes fungi, Nitrosomonas , etc. [ 137 , 138 , 140 , 141 ]. For specific contaminant degradation, predominantly well-defined microorganisms are used.
To improve the potency of proteins to overexpress the desired character for degradation by transforming microbes using genetic engineering approaches where they are transfected with genes that encode catabolic enzymes. Nowadays, genetically engineered microorganisms (GEM) are the most feasible xenobiotic-degrading microorganisms ( E. coli and Pseudomonas putida ) in wastewater treatment, and with the help of GEM, we can improve the bioaugmentation process. These GEMs have been used to degrade hexane, oil spills, xylene, toluene, camphor, trichloroethylene, etc. because of their high degradative capacities for various pollutants in wastewater [ 131 , 142 ]. Manipulation of the oil-degrading Pseudomonas bacterium with plasmids containing genes encoding catabolic enzymes used in the degradation of aromatic compounds [ 143 , 144 ]. For biodegradation of atrazine, metal removal, and direct blue dye in waste water, a genetically modified E. coli strain has been used [ 145 , 146 ].
Genetically modified E. coli involved in wastewater treatment
Mercury (Hg) is the most dangerous heavy metal that can be released into the environment through industrial wastewater. Mercury can be removed from contaminated water, soil, or sediment by the GE E. coli strain JM109 [ 43 ]. Mercury can be removed from a contaminated site using GE bacteria that possess the MerA gene [ 147 , 148 ]. GE E. coli has been discovered to digest trichloroethylene after being transformed with a variety of phenol catabolic genes such as pheA, pheB, pheC, pheD, and pheR. Nickel (Ni) is perhaps the most tenacious toxin, and it can be extracted from water by the GE E. coli SE5000 strain [ 149 ]. In this way, GE microorganisms can help with the bioremediation of heavy metals from degraded sites.
Safety of using recombinant E. coli strain for treatment of pollutants
Artificial generation of pollutants takes place by various by-products produced by the modern human world, which leads to toxicological impacts on nature. With growing awareness about the direct and indirect impacts of environmental pollution on ecosystems, efficient, cost-effective, and environmentally safe methods are being developed for the treatment of pollutants. The rapid rise in the rate of industrialization and the manufacturing of harmful toxic products leads to a change in the homeostatic balance of ecological biodiversity. Recombinant DNA technology emerged in 1972 and became the cutting-edge technology in the modern world, leading to the mass production of human insulin, human growth hormones, interferon, and the hepatitis vaccine [ 150 ]. In medical sciences, delivery made by this technology has set mild stones to combat pollution. For this purpose, genetically modified organisms (GMO) produced by recombinant DNA technology are used as a promising option for the treatment of pollutants, and many reports have also been published in this context [ 151 , 152 ]. There are a variety of pollutants that are increasing at an alarming rate in the environment and need to be monitored.
The common pollutants that are to be taken into consideration are heavy metals, high density petroleum hydrocarbons (mercury, lead, arsenic, cadmium, etc.), polymers, chlorinated hydrocarbons, pesticides, insecticides (polycarbonates, polyethylene, polyurethane, polypropylene, etc.), explosives, detergents (GTN, TNT, and RDX), etc. [ 153 , 154 ]. The combination of biotechnology and recombinant DNA technology is improving pollutant-degrading microbes through genetic modifications and strain improvement of specific metabolic and regulatory genes that are crucial in biodegradation [ 30 ]. Chakrabarty [ 155 ] established the bar by patenting petroleum oil pollution bioremediation, which was the first step towards using recombinant DNA technology for pollution mitigation. The most important and prominent tools for recombinant DNA technology are GMOs, which aid in the bioremediation of pollutants. Although they are potential deliverables, they need statutory clearance to be used in an open environment in many countries. So various regulatory guidelines are framed in different countries for their safe use.
The recombinant E. coli K-12 strain is extensively used for pollution control as it does not colonize the human gut and is non-pathogenic [ 156 ]. Further, it has a simple expression system as compared to other higher-level organisms and a large quantity of well-characterized genomic databases. Although certain scientific considerations are to be taken into account while assessing the environmental use of this recombinant microorganism by selecting appropriate safety measures, this may pose some negative impact on the environment [ 157 ]. The bioremediation process is monitored indirectly by measuring the polluted site’s redox potential as well as temperature, pH, electron acceptor and donor concentrations, oxygen content, and concentrations of breakdown products (e.g., carbon dioxide), and petroleum-contaminated environments are analyzed by bacterial biosensors [ 158 , 159 ]. In addition, microbial biosensors are increasingly being used to detect contaminants in systems based on reporter genes.
The specific metals in cellular environments are responsible for the expression of resistance genes, and this specificity of tight regulation is exploited in such biosensors [ 154 ]. The promoters and regulatory genes present in resistance operons are being used to construct metal-specific biosensors (promoter-reporter gene fusions) [ 160 ]. In addition to chemical analysis, metal-specific biosensors can be used:
To regulate pollutant concentration
Bioavailable metal concentration in the samples [ 110 ]
Thus, currently existing risk assessment and safety methods are being used to characterize the consequences of human exposure to such E. coli strains. Further, key difficulty lies in the assessment of interactions of the microorganism with the existing ecosystem. For example, an introduced E. coli strain may pass genetic material to other microbes, altering the environment and resulting in secondary impacts. Thus, two important areas of investigation related to establishment and proliferation are as follows:
(i) Fate of the recombinant E. coli strain and environmental transfer.
(ii) Interaction with the ecosystem.
This knowledge of recombinant E. coli transport and its fate (or survival) is useful for assessing potential exposures of nontarget organisms or nontarget areas and rendering it safe for remediation of pollutants [ 161 , 162 ].
Policy regarding regulation of genetically modified microorganisms
The recent advancements in genetic manipulation offer vast potential and are being utilized in various innovative experiments and applications. These progressions have raised apprehensions among researchers in the biological sciences and other related fields regarding the safe conduct of research in this domain. Genetically modified organisms (GMOs) and their products are regulated in India under the “Rules for the manufacture, use, import, export & storage of hazardous microorganisms, genetically engineered organisms or cells, 1989” (referred to as Rules, 1989) notified under the Environment (Protection) Act, 1986 [ 163 ]. The Ministry of Environment, Forest, and Climate Change, the Department of Biotechnology, and state governments enforce these rules through six competent authorities. Six competent authorities and their composition have been notified under these rules that include the following: rDNA Advisory Committee (RDAC), Institutional Biosafety Committee (IBSC), Review Committee on Genetic Manipulation (RCGM), Genetic Engineering Appraisal Committee (GEAC), State Biotechnology Coordination committee (SBCC), and District Level Committee (DLC). The Recombinant DNA Advisory Committee (RDAC) has been established by the department for this specific reason. A publication outlining the Recombinant DNA Safety Guidelines has been released, based on the latest scientific knowledge, to regulate the use of this technique in research, production, and various applications. In 2014, the Department of Biotechnology (DBT) established a specialized task force focused on “Genome Engineering Technologies and their Applications.”
The Coordinated Framework for Regulation of Biotechnology was issued in 1986 by the Office of Science and Technology Policy (OSTP) in United States of America (USA). The framework detailed the allocation of regulatory duties among the many authorities that deal with pesticide, food, and agricultural goods. Therefore, in compliance with the framework, the US Environmental Protection Agency (US EPA) regulates microorganisms and other genetically engineered constructs intended for pesticidal purposes and subject to the Federal Insecticide Fungicide and Rodenticide Act (FIFRA) and the Federal Food Drug and Cosmetic Act (FFDCA); USDA APHIS regulates microbes that are plant pests under the Plant Protection Act (PPA) and the National Environmental Policy Act (NEPA). Additionally, certain genetically modified microbes employed as biofertilizers, bioremediation agents, and to produce other industrial compounds including biofuels under the Toxic Substances Control Act (TSCA) are regulated by the US EPA (EPA 1999) [ 164 ].
The European Union (EU) has put in place a number of legal tools to guarantee the safety of goods made with or containing GMMs. A product must undergo a scientific risk assessment before it is allowed to be sold. A guidebook for the risk evaluation of genetically modified organisms (GMOs) in food or feed products has been released by the European Food Safety Authority’s (EFSA) GMO Panel (EFSA, 2011) [ 165 ]. The evaluation is divided into two sections: the GMOs characterization and any potential impact the modification may have on the product’s overall safety.
Genetic engineering methods have provided enough opportunities to remove pollutants and toxins from the environment. Comparing this technology with conventional technologies, it is less expensive and more ecologically friendly. It is important to consider environmental factors that may influence the bioremediation of contaminated sites. Microorganisms have an optimal environment for maximum performance as well as a limit of adaptation to certain environmental conditions. A range of biochemical, microbiological, ecological, and genetic factors influence the rate of bioprocessing and biodegradation of contaminants by genetically engineered bacteria for environmental cleanup. Scientists are continually uncovering new unique genes that can be used to generate new constructs and eventually a new strain that aids in the manufacture of derivative routes for new synthetic compounds, as well as the introduction of biodegradation capabilities in a variety of locations. Even with their great potential and encouraging results in the treatment of pollutants by recombinant host bacteria, recombinant bacteria still face a number of difficulties in the process of treating pollutants. In a complex environment with several substrates and numerous microbial interactions, only a small number of modified bacteria are involved in the treatment and removal of toxins. The greatest way to increase biodegradation variety is to use a plasmid with multiple operons rather than multi-plasmids with favorable qualities as plasmids are not only compatible but also incompatible. Protoplast fusion technique has demonstrated promising outcomes in the breeding of biodegradation-engineered bacteria, in addition to plasmids, which is a good thing. Recombinant bacterial strains were produced using the protoplast fusion process; however, the strains also contained genes that were unnecessary or harmful to breakdown. It is also important to follow the correct regulatory procedures for the safe containment and use of GMOs in bioremediation processes.
The subsequent stages of bioremediation research involve discovering and comparing gene and protein sequences that are efficient at eliminating contaminants, even though genomics, metabolomics, and proteomics in bioremediation help explore potential solutions to particular pollutants. GMOs have the ability to clean up a variety of contaminated soil and waste effluents. Utilizing bioremediation in tandem with other physical and chemical techniques can offer an all-encompassing strategy for eliminating pollutants from the surroundings and has the potential to overcome current challenges. It seems to be a long-term treatment; thus, more study in this field is required.
Availability of data and materials
No datasets were generated or analysed during the current study.
Patel AB, Jain KR, Manvar T, Desai C, Madamwar D. Enriched bacterial community efficiently degrade polycyclic aromatic hydrocarbons in soil ecosystem: insights from a mesocosms study. Biochem Eng J. 2022;185: 108516. https://doi.org/10.1016/j.bej.2022.108516 .
Article CAS Google Scholar
Siddiqua A, Hahladakis JN, Attiya WAKA. An overview of the environmental pollution and health effects associated with waste landfilling and open dumping. Environ Sci Pollut Res Int. 2022;29:58514–36.
Article PubMed PubMed Central Google Scholar
Xu Y, Xue X, Dong L, Nai C, Liu Y, Huang Q. Long-term dynamics of leachate production, leakage from hazardous waste landfill sites and the impact on groundwater quality and human health. Waste Manag. 2018;82:156–66. https://doi.org/10.1016/j.wasman.2018.10.009 .
Article CAS PubMed Google Scholar
Nadal M, Rovira J, Díaz-Ferrero J, Schuhmacher M, Domingo J. Human exposure to environmental pollutants after a tire landfill fire in Spain: health risks. Environ Int. 2016;97:37–44. https://doi.org/10.1016/j.envint.2016.10.016 .
Gworek B, Dmuchowski W, Koda E, Marecka M, Baczewska HA, Brągoszewska P, Sieczka A, Osiński P. Impact of the municipal solid waste Łubna landfill on environmental pollution by heavy metals. Water. 2016;2016(8):470. https://doi.org/10.3390/w8100470 .
Ozkara A, Akyıl D, Konuk M. Pesticides, environmental pollution, and health, In Environmental Health Risk - Hazardous Factors to Living Species. In: Larramendy ML, Soloneski S, editors. CBS Publishers; 2016. https://doi.org/10.5772/63094 .
Ozkara A, Akyil D. Environmental pollution and pollutants of ecosystem: a review. Turkish Journal of Scientific Reviews. 2018;11:11–7.
Google Scholar
Joutey NT, Bahafid W, Sayel H, Ghachtouli NE. Biodegradation: involved microorganisms and genetically engineered microorganisms. Agricultural and Biological Sciences: Biodegradation - Life of Science Editors: Rolando Chamy and Francisca Rosenkranz. 2013. https://doi.org/10.5772/56194 .
Article Google Scholar
Rabani MS, Sharma R, Singh R, Gupta M. Characterization and identification of naphthalene degrading bacteria isolated from petroleum contaminated sites and their possible use in bioremediation. Polycyclic Aromat Compd. 2022;42(3):978–89. https://doi.org/10.1080/10406638.2020.1759663 .
Ghosh D, Parida P. Air pollution and India: current scenario. Internation J Curr Res. 2015;7(11):22194–6.
Ojuederie OB, Babalola OO. Microbial and plant-assisted bioremediation of heavy metal polluted environments: a review. Int J Environ Res Public Health. 2017;14:1504–10. https://doi.org/10.3390/ijerph14121504 .
Article CAS PubMed PubMed Central Google Scholar
Bhatia RK, Sakhuja D, Mundhem S, Walia A. Renewable energy products through bioremediation of wastewater. Sustainability. 2020;12(7501):1–24. https://doi.org/10.3390/su12187501 .
Ekperusi O, Aigbodion F. Bioremediation of petroleum hydrocarbons from crude oil-contaminated soil with the earthworm: Hyperiodrilus africanus. 3 Biotech. 2015;5:957–65. https://doi.org/10.1007/s13205-015-0298-1 .
Ayangbenro AS, Babalola OO. A new strategy for heavy metal polluted environments: a review of microbial biosorbents. Int J Environ Res. 2017;14:94. https://doi.org/10.3390/ijerph14010094 .
Kalia A, Sharma S, Semor N, Babele PK, Sagar S, Bhatia RK, et al. Recent advancements in hydrocarbon bioremediation and future challenges: a review. 3 Biotech. 2022;12(135). https://doi.org/10.1007/s13205-022-03199-y .
Tak HI, Ahmad F, Babalola OO. Advances in the application of plant growth-promoting rhizobacteria in phytoremediation of heavy metals. In: Whitacre DM, editor. Reviews of Environmental Contamination and Toxicology. New York: Springer; 2013;223:33–52. https://doi.org/10.1007/978-1-4614-5577-6_2 .
Ghai H, Sakhuja D, Yadav S, Solanki P, Putatunda C, Bhatia RK, Bhatt AK, Varjani S, Yang YH, Bhatia SK, Walia A. An overview on co-pyrolysis of biodegradable and non-biodegradable wastes. Energies. 2022;15(11):4168. https://doi.org/10.3390/en15114168 .
Bhandari G, Gupta S, Chaudhary P, Chaudhary S, Gangola S. Bioleaching: a sustainable resource recovery strategy for urban mining of e-waste. In: Debbarma P, Kumar S, Suyal DC, Soni R, editors. Microbial technology for sustainable e-waste management. Cham: Springer; 2023. p. 157–75.
Chapter Google Scholar
Shekhar SK, Godheja J, Modi DR. Hydrocarbon bioremediation efficiency by five indigenous bacterial strains isolated from contaminated soils. Intern J Curr Microbiol Appl Sci. 2015;3:892–905.
Pooja N, Chakraborty I, Rahman MH, et al. An insight on sources and biodegradation of bioplastics: a review. 3 Biotech. 2023;13:220. https://doi.org/10.1007/s13205-023-03638-4 .
Kebede G, Tafese T, Abda EM, Kamaraj M, Assefa F. Factors influencing the bacterial bioremediation of hydrocarbon contaminants in the soil: mechanisms and impacts. J Chem. 2021;1–17. https://doi.org/10.1155/2021/9823362 .
Bala S, Garg D, Thirumalesh BV, Sharma M, Sridhar K, Inbaraj BS, et al. Recent strategies for bioremediation of emerging pollutants: a review for a green and sustainable environment. Toxics. 2022;10(8):484. https://doi.org/10.3390/toxics10080484 .
Sharma P, Singh SP, Parakh SK, Tong YW. Health hazards of hexavalent chromium (Cr (VI)) and its microbial reduction. Bioengineered. 2022;13(3):4923–38. https://doi.org/10.1080/21655979.2022.2037273 .
Mahmoud GAE. Microbial scavenging of heavy metals using bioremediation strategies. In: Vivek K, Ram P, Manoj K, editors. Rhizobiont in bioremediation of hazardous waste. Singapore: Springer; 2021. p. 265–89. https://doi.org/10.1007/978-981-16-0602-1_12 .
Ambaye TG, Vaccari M, Franzetti A, Prasad S, Formicola F, Rosatelli A, et al. Microbial electrochemical bioremediation of petroleum hydrocarbons (PHCs) pollution: recent advances and outlook. Chem Eng J. 2023;452:139372. https://doi.org/10.1016/j.cej.2022.139372 .
AL-Huqail AA, Kumar P, Eid EM, Adelodun B, Abou Fayssal S, Singh J, et al. Risk assessment of heavy metals contamination in soil and two rice (Oryza sativa L.) varieties irrigated with paper mill effluent. Agriculture. 2022;12(11):1864. https://doi.org/10.3390/agriculture12111864 .
Ifon EB, Alexis T, Tometin LAS, Suanon F. Metal-contaminated soil remediation: phytoremediation, chemical leaching and electrochemical remediation. Intech Open: Metals in Soil - Contamination and Remediation Publisher; 2019. p. 89–98.
Benjedim S, Romero-Cano LA, Pérez-Cadenas AF, Bautista-Toledo MIB, Lotfi EI M, Carrasco-Marín F. Removal of emerging pollutants present in water using an E. coli biofilm supported onto activated carbons prepared from argan wastes: adsorption studies in batch and fixed bed. Sci Total Environ. 2020;720. https://doi.org/10.1016/j.scitotenv.2020.137491
Zhang J, Zhang H, Li S, Li J, Yan L, Xia L. Increasing yield potential through manipulating of an ARE1 ortholog related to nitrogen use efficiency in wheat by CRISPR/Cas9J. Integr Plant Biol. 2020;63(9):1649–63. https://doi.org/10.1111/jipb.13151 . (Epub 2021 Sep 2).
Yaashikaa PR, Devi MK, Kumar PS. Engineering microbes for enhancing the degradation of environmental pollutants: a detailed review on synthetic biology. Environ Res. 2022;214(1):113868. https://doi.org/10.1016/j.envres.2022.113868 .
Perera IC, Hemamali EH. Genetically modified organisms for bioremediation: current research and advancements. In: Suyal DC, Soni R, editors. Bioremediation of Environmental Pollutants. Cham: Springer; 2022. https://doi.org/10.1007/978-3-030-86169-8_7 .
Sanghvi G, Thanki A, Pandey S and Singh NK. Engineered bacteria for bioremediation. Bioremediation Pollut. 2020;359–74. https://doi.org/10.1016/B978-0-12-819025-8.00017-X .
Chugh M, Kumar L, Shah MP, Bharadvaja N. Algal bioremediation of heavy metals; an insight into removal mechanisms, recovery of by-products, challenges, and future opportunities. Energy Nexus. 2022;7:100129. https://doi.org/10.1016/j.nexus.2022.100129 .
Zhang W, Lin Z, Pang S, Bhatt P, Chen S. Insights into the biodegradation of lindane (γ-hexachlorocyclohexane) using a microbial system. Frontier in Microbiology. 2021;2021(11):522. https://doi.org/10.3389/fmicb.2020.00522 .
Okoli AS, Blix T, Myhr AI, Xu, W, Xu X. Sustainable use of CRISPR/Cas in fish aquaculture: the biosafety perspective. Transgenic Res. 2021:1–21. https://doi.org/10.1007/s11248-021-00274-7 .
Pyne ME, Young MY, Chung DA, Chou CP. Coupling the CRISPR/Cas9 system to lambda red recombineering enables simplified chromosomal gene replacement in Escherichia coli . Appl Environ Microbiol. 2015;81(15):5103–14. https://doi.org/10.1128/AEM.01248-15 .
Gong T, Xu X, Dang Y, Kong A, Wu Y, Liang P, Wang S, Yu H, Xu P, Yang C. An engineered Pseudomonas putida can simultaneously degrade organophosphates, pyrethroids and carbamates. Sci Total Environ. 2018;628:1258–65. https://doi.org/10.1016/j.scitotenv.2018.02.143 .
Gallo G, Puopolo R, Carbonaro M, Maresca E, Fiorentino G. Extremophiles, a nifty tool to face environmental pollution: from exploitation of metabolism to genome engineering. Int J Environ Res Publ Health. 2021;18(10):5228. https://doi.org/10.3390/ijerph18105228 .
Adamczyk PA, Reed JL. Escherichia coli as a model organism for systems metabolic engineering. Curr Opin Sys Biol. 2017;6:80–8.
Ali S, Bukhari DA, Rehmanb A. Call for biotechnological approach to degrade plastic in the era of COVID-19 pandemic. Saudi J Biol Sci. 2023;30(3).
Bala GP, Rajnoveanu RM, Tudorache E, Motisan R, Oancea C. Air pollution exposure–the (in) visible risk factor for respiratory diseases. Environ Sci Pollut Res. 2021;28:19615–28. https://doi.org/10.1007/s11356-021-13208-x .
Burlingame R, Champman PJ. Catabolism of phenyl propionic acid and its 3-hydroxy derivatives by E.coli . J Bacteriol. 1983;155:113–21.
Chen S, Wilson DB. Genetic engineering of bacteria and their potential for Hg 2+ bioremediation. Biodegrad. 1997;8:97–103. https://doi.org/10.1023/a:1008233704719 .
Valls M, Atrian S, de Lorenzo V, Herrero LÁ. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat Biotechnol. 2000;18(6):661–5. https://doi.org/10.1038/76516 .
Varjani S, Rakholiya P, Ng HY, You S, Teixeira JA. Microbial degradation of dyes: an overview. Biores Technol. 2020;314:123728. https://doi.org/10.1016/j.biortech.2020.123728 .
Mohamed MSM, et al. Reduction of chromium-VI by chromium-resistant Escherichia coli FACU: a prospective bacterium for bioremediation. Folia Microbiol. 2020. https://doi.org/10.1007/s12223-020-00771-y .
Bhatt P, Gangola S, Joshi C, Chaudhary P, Kumar G, Bhandari G, et al. Recent advancements and mechanism of microbial enzymes in sustainable agriculture. Microb Technol Sustain Environ. 2021;1:247–59. https://doi.org/10.1007/978-981-16-3840-4_15 .
Bhatt P, Verma A, Gangola S, Bhandari G, Chen S. Microbial glycoconjugates in organic pollutant bioremediation: recent advances and applications. Microb Cell Factories. 2021;20(1):1–18. https://doi.org/10.1186/s12934-021-01556-9 .
Saeed MU, Hussain N, Sumrin A, Shahbaz A, Noor S, Bilal M, et al. Microbial bioremediation strategies with wastewater treatment potentialities – a review. Sci Total Environ. 2022;818. https://doi.org/10.1016/j.scitotenv.2021.151754 .
Chaudhary P, Ahamad L, Chaudhary A, Kumar G, Chen WJ, Chen S. Nanoparticle-mediated bioremediation as a powerful weapon in the removal of environmental pollutants. J Environ Chem Eng. 2023;11:109591. https://doi.org/10.1016/j.jece.2023.109591 .
Menn FM, Easter JP, Sayler G. Genetically engineered microorganisms and bioremediation. In: Rehm H-J, Reed G, editors. Biotechnology: Environmental Processes II 2008; 11b, Second Edition. Weinheim: Wiley-VCH Verlag GmbH; 2008. https://doi.org/10.1002/9783527620951.ch21 .
Xia QJ, Li Y, Xu T, Wu K. Display of lead-binding proteins on Escherichia coli surface for lead bioremediation. Biotechnol Bioeng. 2020;117(12):3820–34. https://doi.org/10.1002/bit.27525 .
Chaudhary P, Xu M, Ahamad L, Chaudhary A, Kumar G, Adeleke BS, et al. Application of synthetic consortia for improvement of soil fertility, pollution remediation, and agricultural productivity: a review. Agronomy. 2023;13(3):643. https://doi.org/10.3390/agronomy13030643 .
Dave S, Das J. Role of microbial enzymes for biodegradation and bioremediation of environmental pollutants: challenges and future prospects. Bioremediat Environ Sustain. 2021;325–46. https://doi.org/10.1016/B978-0-12-820524-2.00013-4 .
He S, Zhang Z, Lu W. Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol. 2023;50(1):kuac029. https://doi.org/10.1093/jimb/kuac029 .
French KE, Zhongrui Z, Terry N. Horizontal ‘gene drives’ harness indigenous bacteria for bioremediation. Sci Reports. 2020;10(1):15091.
CAS Google Scholar
Xia X, Wu S, Zhou Z, Wang G. Microbial Cd (II) and Cr (VI) resistance mechanisms and application in bioremediation. J Hazard Mater. 2021;401:123685. https://doi.org/10.1016/j.jhazmat.2020.123685 .
Lu CW, Ho HC, Yao CL, Tseng TY, Kao CM, Chen SC. Bioremediation potential of cadmium by recombinant Escherichia coli surface expressing metallothionein MTT5 from Tetrahymena thermophila. Chemosphere. 2023;310. https://doi.org/10.1016/j.chemosphere.2022.136850 .
Vulpe CB, Matica MA, Kovacevic R, Dascalu D, Stevanovic Z, Isvoran A, Ostafe V, Menghiu G. Copper accumulation efficiency in different recombinant microorganism strains available for bioremediation of heavy metal-polluted waters. Int J Mol Sci. 2023;24(8). https://doi.org/10.3390/ijms24087575 .
Deng X, Li QB, Lu YH, He N, Jiang J. Genetic engineering of E. coli SE5000 and its potential for Ni bioremediation. Process Biochem. 2005;40(1):425–30. https://doi.org/10.1016/j.procbio.2004.01.019 .
Kao WC, Wu JY, Chang CC, Chang JS. Cadmium biosorption by polyvinyl alcohol immobilized recombinant Escherichia coli. J Hazard Mater. 2009;169(3):651–8. https://doi.org/10.1016/j.jhazmat.2009.03.140 .
Tsyganov VE, et al. Efficacy of a plant-microbe system: Pisum sativum (L.) cadmium-tolerant mutant and Rhizobium leguminosarum strains, expressing pea metallothionein genes PsMT1 and PsMT2, for cadmium phytoremediation. Front Microbiol. 2020;11:15. https://doi.org/10.3389/fmicb.2020.00015 .
Strong LC, McTavish H, Sadowsky MJ, et al. Field-scale remediation of atrazine-contaminated soil using recombinant Escherichia coli expressing atrazine chlorohydrolase. Environ Microbiol. 2000;2(1):91–8. https://doi.org/10.1046/j.1462-2920.2000.00079.x .
Zhao XW, Zhou MH, Li QB, Lu Y, He N, Sun DH, Deng X. Simultaneous mercury bioaccumulation and cell propagation by genetically engineered Escherichia coli. Process Biochem. 2005;40(5):1611–16. https://doi.org/10.1016/j.procbio.2004.06.014 .
Yang J, Liu R, Song W, Yang Y, Cui F, Qiao C. Construction of a genetically engineered microorganism that simultaneously degrades organochlorine and organophosphate pesticides. Appl Biochem Biotechnol. 2012;166(3):590–8. https://doi.org/10.1007/s12010-011-9450-5 .
Xiaoqiang J, et al. Display of lead-binding proteins on Escherichia coli surface for lead bioremediation. Biotechnol Bioeng. 2020;117(12):3820–34. https://doi.org/10.1002/bit.27525 .
Dellanno F, Rastelli E, Tangherlini M, Corinaldesi C, Sansone C, Brunet C, et al. Highly contaminated marine sediments can host rare bacterial taxa potentially useful for bioremediation. Front Microbiol. 2021;12:584850. https://doi.org/10.3389/fmicb.2021.584850 .
Article PubMed Google Scholar
Cachada A, Rocha-Santos T, Duarte AC. Soil and pollution: an introduction to the main issues. In Soil pollution. Academic Press. p. 1–28. https://doi.org/10.1016/B978-0-12-849873-6.00001-7 .
Dhaka A, Chattopadhyay P. A review on physical remediation techniques for treatment of marine oil spills. J Environ Manage. 2021;288:112428. https://doi.org/10.1016/j.jenvman.2021.112428 .
Duc H, Hung N, Oanh N. Anaerobic degradation of endosulfans by a mixed culture of Pseudomonas sp. and Staphylococcus sp. Appl Biochem Microbiol. 2021;57(3):327–34. https://doi.org/10.1134/S0003683821030030 .
Saha JK, Selladurai R, Coumar MV, Dotaniya ML, Kundu S, Patra AK. Soil pollution - an emerging threat to agriculture. Environ Chem Sustainable World. 2017;10:386.
Duarte A, Cachada A, Rocha-Santos T. Soil pollution: from monitoring to remediation soil and pollution: an introduction to the main issues. Elsevier Inc.; 2017. https://doi.org/10.1111/sum.12443 .
Dutta N, Usman M, Ashraf MA, Luo G, Zhang S. Efficiency of emerging technologies in addressing reductive dechlorination for environmental bioremediation: a review. J Hazard Mater Lett. 2022;3:100065. https://doi.org/10.1016/j.hazl.2022.100065 .
Mishra A, Kumari M, Kumar R, Iqbal K, Thakur IS. Persistent organic pollutants in the environment: risk assessment, hazards, and mitigation strategies. Bioresour Technol Rep. 2022;19. https://doi.org/10.1016/j.biteb.2022.101143 .
Gaur VK, Tripathi V and Manickam N. Bacterial- and fungal-mediated biodegradation of petroleum hydrocarbons in soil. Development in wastewater treatment research and processes. Amsterdam: Elsevier; 2022. p. 407–27. https://doi.org/10.1016/B978-0-323-85839-7.00008-6 .
Azhar U, Ahmad H, Shafqat H, Babar M, Munir HMS, Sagir M, Arif M, Hassan A, Rachmadona N, Rajendran S, Mubashir M, Khoo KS. Remediation techniques for elimination of heavy metal pollutants from soil: a review. Environ Res. 2022;214(4). https://doi.org/10.1016/j.envres.2022.113918 .
Garbisu C, Garaiyurrebaso O, Epelde L, Grohmann E. Plasmid-mediated bioaugmentation for the bioremediation of contaminated soils. Front Microbiol. 2017;8:1–13. https://doi.org/10.3389/fmicb.2017.01966 .
Smarzewska S, Guziejewski D. Soil remediation technologies. In: Rehab O. Abdel Rahman, Chaudhery Mustansar Hussain, editors. Handbook of Advanced Approaches Towards Pollution Prevention and Control. Elsevier; 2021. p. 193–219. https://doi.org/10.1016/B978-0-12-822121-1.00010-2 .
Ashraf MA, Maah MJ, Yusoff I. Soil contamination, risk assessment and remediation. In: Environmental Risk Assessment of Soil Contamination. London: IntechOpen; 2014. p. 1–56. https://doi.org/10.5772/57287 .
Almaguer-Cantú V, Morales-Ramos Lilia H and Balderas-Rentería I. Biosorption of lead (II) and cadmium (II) using Escherichia coli genetically engineered with mice metallothionein I. Water Sci Technol. 2011;63(8):1607–13. https://doi.org/10.2166/wst.2011.225 .
Geetha N, Bhavya G, Abhijith P, Shekhar R, Dayananda K, Jogaiah S. Insights into nanomycoremediation: secretomics and mycogenic biopolymer nanocomposites for heavy metal detoxification. J Hazard Mate. 2021;409:124541. https://doi.org/10.1016/j.jhazmat.2020.124541 .
Priyadarshanee M, Das S. Biosorption and removal of toxic heavy metals by metal tolerating bacteria for bioremediation of metal contamination: a comprehensive review. J Environ Chem Eng. 2021;9(1):2213–3437. https://doi.org/10.1016/j.jece.2020.104686 .
Pham VHT, Kim J, Chang S, Chung W. Bacterial biosorbents, an efficient heavy metals green clean-up strategy: prospects, challenges, and opportunities. Microorganisms. 2022;10(3):610. https://doi.org/10.3390/microorganisms10030610 .
Hitt LG, Khalil S, Blanchette A, Finkelstein ME, Iverson EN, McClellannd SC, et al. Lead exposure is correlated with reduced nesting success of an urban songbird. Environ Res. 2023;227(1):115711. https://doi.org/10.1016/j.envres.2023.115711 .
Sriprang R, et al. A novel bioremediation system for heavy metals using the symbiosis between leguminous plant and genetically engineered rhizobia. J Biotechnol. 2002;99(3):279–93. https://doi.org/10.1016/S0168-1656(02)00219-5 .
Wu WM, Carley J, Fienen M, Mehlhorn T, Lowe K, Nyman J, et al. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer. 1. Conditioning of a treatment zone. Environ Sci Technol. 2006;40:3978–85. https://doi.org/10.1021/es051954y .
Bondarenko O, Rõlova T, Kahru A, Ivask A. Bioavailability of Cd, Zn and Hg in soil to nine recombinant luminescent metal sensor bacteria. Sensors. 2009;8(11):6899–923. https://doi.org/10.3390/s8116899 .
Ivask A, Dubourguier HC, Põllumaa L, Kahru A. Bioavailability of Cd in 110 polluted top soils to recombinant bioluminescent sensor bacteria: effect of soil particulate matter. J Soils Sedim. 2011;11(2):231–7. https://doi.org/10.1007/s11368-010-0292-5 .
Liu PWG, et al. Bioremediation of petroleum hydrocarbon contaminated soil: effects of strategies and microbial community shift. Int Biodeter Biodegr. 2011;65(8):1119–27. https://doi.org/10.1016/j.ibiod.2011.09.002 .
Bae W, Wu CH, Kostal J, Mulchandani A, Chen W. Enhanced mercury biosorption by bacterial cells with surface-displayed MerR. Appl Environ Microbiol. 2003;69(6):3176–80. https://doi.org/10.1128/AEM.69.6.3176-3180.2003 .
Kiyono M, Pan-Hou H. Genetic engineering of bacteria for environmental remediation of mercury. J Health Sci. 2006;52(3):199–204. https://doi.org/10.1248/jhs.52.199 .
Vu KA, Mulligan CN. An overview on the treatment of oil pollutants in soil using synthetic and biological surfactant foam and nanoparticles. Int J Mol Sci. 2023;24(3):1916. https://doi.org/10.3390/ijms24031916 .
Gricajeva A, Nadda AK, Gudiukaite R. Insights into polyester plastic biodegradation by carboxyl ester hydrolases. J Chem Technol Biotechnol. 2022;97(2):359–80. https://doi.org/10.1002/jctb.6745 .
Hussain A, Rehman F, Rafeeq H, Waqas M, Asghar A, Afsheen N, et al. In-situ, ex-situ, and nano-remediation strategies to treat polluted soil, water, and air-a review. Chemosphere. 2022;289:133252. https://doi.org/10.1016/j.chemosphere.2021.133252 .
Kumar A, Sharma A, Chaudhary P, Gangola S. 2021. Chlopyrifos degradation using binary fungal strains isolated from industrial waste soil. Biologia. 2021;76(10):3071–83. https://doi.org/10.1007/s11756-021-00816-8 .
Liu L, Bilal M, Duan X, Iqbal HMN. Mitigation of environmental pollution by genetically engineered bacteria — current challenges and future perspectives. Sci Total Environ. 2019;667:444–54. https://doi.org/10.1016/j.scitotenv.2019.02.390 .
Punyamurthy C, Bheenaveni RS. Urbanization in India: an overview of trends causes and challenges. Int J Asian Econ Light. 2023;11(1). https://doi.org/10.36713/epra12473 .
Ng NL, Brown SS, et al. Nitrate radicals and biogenic volatile organic compounds: oxidation, mechanisms, and organic aerosol. Atmos Chem Physics. 2017;17:2103. https://doi.org/10.5194/acp-17-2103-2017 .
David E, Niculescu VC. Volatile organic compounds (VOCs) as environmental pollutants: occurrence and mitigation using nanomaterials. Int J Environ Res Public Health. 2021;18(24):13147. https://doi.org/10.3390/ijerph182413147 .
D’Amato G, Pawankar R, Vitale C, Maurizia L. Climate change and air pollution: effects on respiratory allergy. Allergy Asthma Immunol Res. 2016;8:391–5. https://doi.org/10.4168/aair.2016.8.5.391 .
Xu M, Wu M, Zhang Y, et al. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by bacterial mixture. Int J Environ Sci Technol. 2022;19:3833–44. https://doi.org/10.1007/s13762-021-03284-4 .
Sathe BM, Khandaskar Y, et al. Chemical composition and source attribution of PM 2.5 and PM 10 in Delhi-National Capital Region (NCR) of India: results from an extensive seasonal campaign. J Atmos Chem. 2021;78:35–58. https://doi.org/10.1007/s10874-020-09412-7 .
Rao MA, Scelza R, Scotti R, Gianfreda L. Role of enzymes in the remediation of polluted environments. J Soil Sci Plant Nutr. 2010;10(3):333–53. https://doi.org/10.4067/S0718-95162010000100008 .
Orlovic-Leko P, Farkas B, Galic I. A short review of environmental and health impacts of gold mining. Reliability. Theory Appl. 2022;17:242–8.
Gupte A, Tripathi A, Patel H, Rudakiya D, Gupte S. Bioremediation of polycyclic aromatic hydrocarbon (PAHs): a perspective. Open Biotechnol J. 2016;2016(10):363–78. https://doi.org/10.2174/1874070701610010363 .
Valavanidis A, Vlachogianni T, Fiotakis K. Air pollution as a significant cause of diseases and premature death: air pollution in urban areas and indoor air pollution are associated with adverse health effects and premature mortality. 2016. WebSite: www.chem-tox-ecotox.org .
Elufisan Temidayo O, Rodríguez-Luna IC, Oyedara OO, Sánchez-Varela A, Hernández-Mendoza A, Gonzalez ED, Paz-González AD, Muhammad K, Rivera G, Villalobos-Lopez MA and Guo X. The polycyclic aromatic hydrocarbon (PAH) degradation activities and genome analysis of a novel strain Stenotrophomonas sp. Pemsol isolated from Mexico. PeerJ. 2020;8:8102. https://doi.org/10.7717/peerj.8102
Nguyen PM, Do PT, Pham YB, Doan TO, Nguyen XC, Lee WK, Nguyen DD, Vadiveloo A, Myoung-Jin, Um Ngo HH. Roles, mechanism of action, and potential applications of sulfur-oxidizing bacteria for environmental bioremediation. Sci Total Environ. 2022;852. https://doi.org/10.1016/j.scitotenv.2022.158203 .
Pokoma D, Zabranska J. Sulfur-oxidizing bacteria in environmental technology. Biotechnol Adv. 2015;33:1246–59. https://doi.org/10.1016/j.biotechadv.2015.02.007 .
Sun M, Andreassi JL, Liu S, Pinto R, Triccas JA, Leyh TS. The trifunctional sulfate-activating complex (SAC) of Mycobacterium tuberculosis. J Biol Chem. 2005;280(9):7861–6. https://doi.org/10.1074/jbc.M409613200 .
Günal S, Hardman R, Kopriva S, Mueller JW. Sulfation pathways from red to green. J Biol Chem. 2019;294(33):12293–312. https://doi.org/10.1074/jbc.REV119.007422 .
Rennenberg H. The fate of excess sulfur in higher plants. Annu Rev Plant Physiol. 1984;35:121–53. https://doi.org/10.1146/annurev.pp.35.060184.001005 .
Cho O, Choi KY, Zylstra GJ, Kim YS, Kim SK, Lee JH, Kim E. Catabolic role of a three-component salicylate oxygenase from Sphingomonas yanoikuyae B1 in polycyclic aromatic hydrocarbon degradation. Biochem Biophys Res Commun. 2005;327(3):656–62. https://doi.org/10.1016/j.bbrc.2004.12.060 .
Jin R, Yang H, Zhang A, Wang J, Liu G. Bioaugmentation on decolorization of CI Direct Blue 71 by using genetically engineered strain Escherichia coli JM109 (pGEX-AZR). J Hazard Mater. 2009;163(2–3):1123–8. https://doi.org/10.1016/j.jhazmat.2008.07.067 .
Khatun R. Water pollution: causes, consequences, prevention method and role of WBPHED with special reference from Murshidabad district. Inter J Scientific Res. 2017;7:269–77.
Khan WA, Khan WA, Ali S, Shah SA. Water pollution: sources and its impact on human health, control and managing. J Int Coop Dev. 2022;5(1):69. https://doi.org/10.36941/jicd-2022-0005 .
Mian IA, Begum S, Riaz M, Ridealgh M, McClean CJ, Cresser MS. Spatial and temporal trends in nitrate concentrations in the River Derwent, North Yorkshire, and its need for NVZ status. Sci Total Env. 2010;408:702–12. https://doi.org/10.1016/j.scitotenv.2009.11.020 .
Jadeja NB, Banerji T, Kapley A, Kumar R. Water pollution in India – current scenario. Water Security. 2022;16:100119. https://doi.org/10.1016/j.wasec.2022.100119 .
Singh N, Poonia T, Siwal SS, Srivastav AL, Sharma HK, Mittal SK. Challenges of water contamination in urban areas. In: Arun Lal Srivastav Sughosh Madhav, Abhishek Kumar Bhardwaj, Eugenia Valsami-Jones, editors. Current Directions in Water Scarcity Research. vol. 6. Elsevier; 2022. p. 173–202. https://doi.org/10.1016/B978-0-323-91838-1.00008 .
Manisalidis I, Stavropoulou E, Stavropoulos A, Bezirtzoglou E. Environmental and health impacts of air pollution: a review front public health. Front Pub Health. 2020;8:14. https://doi.org/10.3389/fpubh.2020.00014 .
Wang D, Li G, Qin S, Tao W, Gong S, Wang J. Remediation of Cr (VI)-contaminated soil using combined chemical leaching and reduction techniques based on hexavalent chromium speciation. Ecotoxicol Environ Saf. 2021;208. https://doi.org/10.1016/j.ecoenv.2020.111734 .
Kumar G, Lal S, Soni SK, Maurya SK, Shukla PK, Chaudhary P, et al. Mechanism and kinectics of chloropyrifos co-metabolism by using environment restoring microbes isolated from rhizosphere of horticultural crops under subtropics. Front Microbiology. 2022;13:2796. https://doi.org/10.3389/fmicb.2022.891870 .
Bhargava A. Wet scrubbers-design of spray tower to control air pollutants. Int J Environ Plann Dev. 2016;2(1):68–73. https://doi.org/10.37628/jepd.v1i1.78 .
Trifunovic V. Vitrification as a method of soil remediation. Zastita Materijala. 2021;62(3):166–79. https://doi.org/10.5937/zasmat2103166T .
Kuruppathparambil RR, Babu R, Jeong HM, Hwang GY, Jeong GS, Kim MI, Kim DW, Park DW. A solid solution zeolitic imidazolate framework as a room temperature efficient catalyst for the chemical fixation of CO 2 . Green Chem. 2016;18(23):6349–56. https://doi.org/10.1039/C6GC01614F .
Koul B, Taak P. Chemical methods of soil remediation. Biotechnological strategies for effective remediation of polluted soils. Singapore: Springer; 2018. https://doi.org/10.1007/978-981-13-2420-8_4
Mousset E, Trellu C, Oturan N, Rodrigo MA, Oturan MA. Soil remediation by electro Fenton process. In: Electro-Fenton process. Singapore: Springer; 2017. p. 399–423. https://doi.org/10.1007/698_2017_38 .
Fayomi G, Mini SE, Fayomi OSI, Owodolu T, Ayoola A, Wusu O. A mini review on the impact of sewage disposal on environment and ecosystem. IOP Conf Ser Earth Environ Sci. 2019;331(1). https://doi.org/10.1088/1755-1315/331/1/012040 .
Karande SA, Thakare SW, Wankhede P, Sakharkar AV. Automatic garbage collector machine. International Journal for Scientific Research and Development. 2018;6(1):1382–4.
Ramanathan S, Sudharshan R. Sewage cleaning machine. 2019. International Journal of Research and Analytical Review. 2019;6(3):124–9.
Kumar V, Bansal V, Madhavan A, Kumar M, Sindhu R, Awasthi MK, Binod P, Saran S. Active pharmaceutical ingredient (API) chemicals: a critical review of current biotechnological approaches. Bioengineered. 2022a;13(2):4309–27. https://doi.org/10.1080/21655979.2022.2031412 .
Pangestu NL, Zahra NL, Sarwono A, Suryawan IWK. Produced water treatment planning using corrugated plate interceptor and ultra filtration for water recycling. Serambi Engineering. 2022;2016(4):2286–93.
Ali SM, Aziz SQ. Dissolved air flotation (DAF) operational parameters and limitations for wastewaters treatment with cost study. Recycling and Sustainable Development. 2023;16:91–7. https://doi.org/10.5937/ror2301091A .
Fairbairn DJ, Trojan MD. Iron-enhanced sand filters: multi-year urban runoff (stormwater) quality performance. Sci Total Environ. 2023;859. https://doi.org/10.1016/j.scitotenv.2022.160177 .
Srinivasan NR, Kamaraj M, Prabhu SV. Strategies and limitations of water treatment methods for point-of-use application. Strategies and tools for pollutant mitigation. 2021. p. 117–133. https://doi.org/10.1007/978-3-030-63575-6_6 .
Saleh IA, Zouari N, Ghouti MAA. Removal of pesticides from water and wastewater: chemical, physical and biological treatment approaches. Environ Technol Innov. 2020;19:101026. https://doi.org/10.1016/j.eti.2020.101026 .
Wan CY, De Wever H, Diels L, et al. Biodiversity and population dynamics of microorganisms in a full-scale membrane bioreactor for municipal wastewater treatment. Water Res. 2011;45:1129–38. https://doi.org/10.1016/j.watres.2010.11.008 .
Hu M, Wang X, Wen X, et al. Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresour Technol. 2012;117:72–9. https://doi.org/10.1016/j.biortech.2012.04.061 .
http://neoakruthi.com/blog/biological-treatment-of-wastewater.html .
Wang J, Liu GF, Lu H, Jin RF, Zhou JT, Lei TM. Biodegradation of acid orange 7 and its auto-oxidative decolorization product in membrane-aerated biofilm reactor. Int Biodeterior Biodegrad. 2012;67:73–7. https://doi.org/10.1016/j.ibiod.2011.12.003 .
McIllroy S, Saunders AM, Albertsen M, et al. MiDAS: the field guide to the microbes of activated sludge. Database (Oxford). 2015. https://doi.org/10.1093/database/bav062) .
Pant G, Garlapti D, Agrawal U, Prasuna RG, Mathimani, Pugazhendhi T. Biological approaches practiced using genetically engineered microbes for a sustainable environment. J Hazard Mat. 2021;405:124631. https://doi.org/10.1016/j.jhazmat.2020.124631 .
McClure NC, Fry JC, Weightman AJ, Survival and catabolic activity of natural and genetically engineered bacteria in a laboratory-scale activated-sludge unit. Appl Environ Microbiol. 1991;57:366–73. https://doi.org/10.1128/aem.57.2.366-373.1991 .
Nusslein K, Maris D, Timmis K, Dwyer DF. Expression and transfer of engineered catabolic pathways harbored by Pseudomonas spp. introduced into activated sludge microcosms. Appl. Environ Microbiol. 1992;58:3380–6. https://doi.org/10.1128/aem.58.10.3380-3386.1992 .
Wei MJ, Wang H, Liu C, Ning DL. Bioaugmentation with immobilized genetically engineered microorganism (GEM)/CAS process for treatment of atrazine wastewater. Huan Jing KeXue. 2008;29:1555–60.
Jan AT, Azam M, Ali A, Haq QMR. Prospects for exploiting bacteria for bioremediation of metal pollution. Crit Rev Environ Sci Technol. 2014;44:519–60. https://doi.org/10.1080/10643389.2012.728811 .
Barkay T, Miller SM, Summers AO. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev. 2005;27:355–84. https://doi.org/10.1016/S0168-6445(03)00046-9 .
De J, Sarkar A, Rahman NS. Bioremediation of toxic substances by mercury resistant marine bacteria. Ecotoxicol. 2006;15:385–9. https://doi.org/10.1007/s10646-006-0066-4 .
Mason JR, Briganti F, Wild JR. Protein engineering for improved biodegradation of recalcitrant pollutants. In: Wild JR, et al, editors. Perspectives in bioremediation. Dordrecht: Kluwer Academic; 1997. p. 107–118. https://doi.org/10.1007/978-90-481-3678-0_8 .
Steinberg FM, Raso J. Biotech pharmaceuticals and biotherapy: an overview. J Pharm Pharmaceutical Sci. 1998;1(2):48–59 (PMID: 10945918).
Glick BR, Pasternak JJ. Molecular biotechnology: principles and applications of recombinant DNA. 3rd Ed. 860. ASM Press; 2002.
Cowan DA. Microbial genomes-the untapped resource. Trends. Biotech. 2000;18:14–6. https://doi.org/10.1016/s0167-7799(99)01395-5 .
Balali-Mood M, Naseri K, Tahergorabi Z, Khazdair MR, Sadeghi M. Toxic mechanisms of five heavy metals: mercury, lead, chromium, cadmium, and arsenic. Front Pharmacol. 2021;12:643972. https://doi.org/10.3389/fphar.2021.643972 .
Frei A, Verderosa AD, Elliott AG, et al. Metals to combat antimicrobial resistance. Nat Rev Chem. 2023;7:202–24. https://doi.org/10.1038/s41570-023-00463-4 .
Chakrabarty AM. Genetically manipulated microorganisms and their products in the oil service Industries. Trends Biotech. 1985;3:32–8. https://doi.org/10.1016/0167-7799(85)90056-3 .
Kelpšas V, Wachenfeldt CV. Strain improvement of Escherichia coli K-12 for recombinant production of deuterated proteins. Sci Rep. 2019;9:17694. https://doi.org/10.1038/s41598-019-54196-w .
Foster-Nyarko E, Pallen MJ. The microbial ecology of Escherichia coli in the vertebrate gut. FEMS Microbiol Rev. 2022;46(3):fuac008. https://doi.org/10.1093/femsre/fuac008 .
Kumar P, Baul G. Recombinant DNA technology for bioremediation of pollutants. In: Fulekar MH, editor. Bioremediation Technology. Dordrecht: Springer; 2010. https://doi.org/10.1007/978-90-481-3678-0_8 .
Huang CW, Lin C, Nguyen MK, Hussain A, Bui XT, Ngo HH. A review of biosensor for environmental monitoring: principle, application, and corresponding achievement of sustainable development goals. Bioengineered. 2023;14(1):58–80. https://doi.org/10.1080/21655979.2022.2095089 .
Misra S, Ganesan M. The impact of inducible promoters in transgenic plant production and crop improvement. Plant Genetics. 2021;27:100300. https://doi.org/10.1016/j.plgene.2021.100300 .
Yu D, Banting G, Neumann Norman F. A review of the taxonomy, genetics, and biology of the genus Escherichia and the type species Escherichia coli. Can J Microbiol. 2021;67(8):553–71. https://doi.org/10.1139/cjm-2020-0508 .
Zhao Q, Yunz S, Bilal M, Hu H. Comparative genomic analysis of 26 Sphingomonas and Sphingobium strains: dissemination of bioremediation capabilities, biodegradation potential and horizontal gene transfer. Sci Total Environ. 2017;609:1238–47.
The Environment (Protection) Act, 1986 vide Notification No. G.S.R. 1198(E) dated 12–11–86 published in the Gazette of India No. 525 dated 12–11–86.
EPA (1999), Status report: TSCA biotechnology submissions. Status report for fiscal years 87–97. http://www.epa.gov/oppt/biotech/pubs/pdf/bistat99.pdf . Accessed 24 June 2011
EFSA. Guidance on the risk assessment of genetically modified microorganisms and their products intended for food and feed use. EFSA J. 2011;9(6):2193.
Goodwin L, Carra I, Campo P, Soares A. Treatment options for reclaiming wastewater produced by the pesticide industry. Int J Water Wastewater Treat. 2017;4(1). https://doi.org/10.16966/2381-5299.149 .
Sharma S, Guleria S, Behl A, Batra N. Isolation and partial characterization of surface producing bacterial strain producing amylase from soil. International Journal of Bio-resources and Stress Management. 2018;9(1):154–8.
Afshari A, Ekberg LE, Forejit L, Mo J, Ardkap SR. Electrostatic precipitators as an indoor air cleaner-a literature review. Sustainability. 2020;12(21):8774. https://doi.org/10.3390/su12218774 .
Ladwig KJ, Blythe GM. Flue-gas desulfurization products and other air emissions controls. Coal Combustion Products. 2017. https://doi.org/10.1016/B978-0-08-100945-1.00003-4 .
Download references
Acknowledgements
I acknowledge the Department of Microbiology, CSK Himachal Pradesh Agricultural University, Palampur, for providing necessary funding to carry out this work.
Author information
Authors and affiliations.
University Institute of Biotechnology, Chandigarh University, Mohali, Punjab, India
Samriti Sharma
Department of Microbiology, Dr. YS Parmar University of Horticulture and Forestry, Nauni, Solan, HP, India
Shruti Pathania & Neha Kaushal
Department of Biotechnology, Jaypee University of Information Technology, Waknaghat, HP, India
Suhani Bhagta
Department of Microbiology, College of Basic Sciences, CSK Himachal Pradesh Agricultural University, Palampur, HP, India
Shivani Bhardwaj & Abhishek Walia
Department of Biotechnology, Himachal Pradesh University, Summer Hill, Shimla, HP, India
Ravi Kant Bhatia
You can also search for this author in PubMed Google Scholar
Contributions
S.S. and S.P. wrote the main Manuscript text. S.B and R.K. prepared figures. S.B and NK prepared Tables. A.W. edit and conceptualized the whole manuscript. All authors reviewed the manuscript.
Corresponding author
Correspondence to Abhishek Walia .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Sharma, S., Pathania, S., Bhagta, S. et al. Microbial remediation of polluted environment by using recombinant E. coli : a review. Biotechnol Environ 1 , 8 (2024). https://doi.org/10.1186/s44314-024-00008-z
Download citation
Received : 08 March 2024
Accepted : 17 May 2024
Published : 13 August 2024
DOI : https://doi.org/10.1186/s44314-024-00008-z
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Genetic engineering
- Bioaccumulation
- Bioaugmentation
Biotechnology for the Environment
ISSN: 2948-2356
- General enquiries: [email protected]
Advertisement
Bioremediation: an emerging effective approach towards environment restoration
- Published: 28 February 2020
- Volume 3 , pages 91–103, ( 2020 )
Cite this article

- Veni Pande 1 , 2 ,
- Satish Chandra Pandey 1 , 2 ,
- Diksha Sati 1 ,
- Veena Pande 2 &
- Mukesh Samant ORCID: orcid.org/0000-0002-0154-2421 1
2311 Accesses
86 Citations
Explore all metrics
Environmental pollution and its remediation are one of the major problems around the globe. Broad varieties of pollutants viz. pesticides, hydrocarbons, heavy metals, and dyes, etc. are the key players, which are mainly responsible for environmental pollution. Residual contaminants are also difficult to eliminate. Bioremediation is one of the most efficient technologies for the reduction of environmental pollutants that recovers the contaminated site back to its actual form. So far only a small number of microbes (culturable microbes) have been exploited and a huge microbial diversity is still unexplored. To enhance the metabolic potential of the microbes, ecological restoration and degradation of recalcitrant pollutants, various bioremediation approaches like chemotaxis, biostimulation, bioaugmentation, biofilm formation, application of genetically engineered microorganisms, advanced omics, have been widely used. In the last few years, the metabolic potential of microbes has tremendously improved the realization of degradation and remediation of environmental pollution. Microorganisms help in the restoration of contaminated habitats by cleaning up waste in a environmentally safe manner along with the production of safe end products. This review discusses the important processes involved in enhancing bioremediation and recent advances in microbes and plants associated bioremediation.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
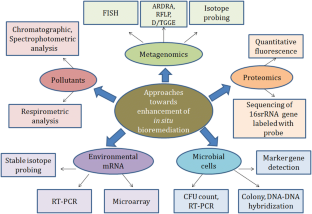
Similar content being viewed by others

Bioremediation: Remedy for Emerging Environmental Pollutants

Microbes: A Potential Tool for Bioremediation

Microbes Assisted Bioremediation: A Green Technology to Remediate Pollutants
Adamson DT, McDade JM, Hughes JB (2003) Inoculation of a DNAPL source zone to initiate reductive dechlorination of PCE. Environ Sci Technol 37:2525–2533
Article CAS Google Scholar
Akilandeswari K, Sona V (2013) Efficiency of Staphylococcus aureusin the degradation an organo phosphorous pesticide Malathion. J Pharm Sci Innov 2:15
Alexander M (1994) Biodegradation and bioremediation. Academic Press, New York
Google Scholar
Alkorta I, Hernández-Allica J, Becerril JM, Amezaga I, Albizu I, Garbisu C (2004) Recent findings on the phytoremediation of soils contaminated with environmentally toxic heavy metals and metalloids such as zinc, cadmium, lead, and arsenic. Rev Environ Sci Biotechnol 3:71–90
Arora NK (2018) Bioremediation: a green approach for restoration of polluted ecosystems. Env Sustain 1:305–307
Article Google Scholar
Arora NK, Panosyan H (2019) Extremophiles: applications and roles in environmental sustainability. Env Sustain 2:217–218
Arora PK, Sasikala C, Ramana CV (2012) Degradation of chlorinated nitroaromatic compounds. Appl Microbiol Biotechnol 93:2265–2277
Ayotamuno JM, Kogbara RB, Agele EA, Agoro OS (2010) Composting and phytoremediation treatment of petroleum sludge. Soil Sediment Contam 19:686–695
Bargiela R, Herbst FA, Martínez-Martínez M, Seifert J, Rojo D, Cappello S (2015) Metaproteomics and metabolomics analyses of chronically petroleum-polluted sites reveal the importance of general anaerobic processes uncoupled with degradation. Proteomics 15:3508–3520
Basumatary B, Bordoloi S, Sarma HP (2012) Crude oil-contaminated soil phytoremediation by using Cyperus brevifolius (Rottb.) Hassk Water. Air Soil Pollut 223:3373–3383
Basumatary B, Saikia R, Chandra Das H, Bordoloi S (2013) Field note: phytoremediation of petroleum sludge contaminated field using sedge species, Cyperus rotundus (Linn.) and Cyperus brevifolius (Rottb.) Hassk. Int J Phytoremediation 15:877–888
Brune KD, Bayer TS (2012) Engineering microbial consortia to enhance biomining and bioremediation. Front Microbiol 2:203
Bursle E, Robson J (2016) Non-culture methods for detecting infection. Aust Prescr 39:171
Chakraborty R, Wu CH, Hazen TC (2012) Systems biology approach to bioremediation. Curr Opin Biotechnol 23:483–490
Chiuchiolo AL (2004) Persistent organic pollutants at the base of the Antarctic marine food web. Environ Sci Technol 38:3551–3557
Cybulski Z, Dzuirla E, Kaczorek E, Olszanowski A (2003) The influence of emulsifiers on hydrocarbon biodegradation by Pseudomonadacea and Bacillacea strains. Spill Sci Technol Bull 8:503–507
Da Silva ML, Alvarez PJ (2004) Enhanced anaerobic biodegradation of benzene-toluene-ethylbenzene-xylene-ethanol mixtures in bioaugmented aquifer columns. Appl Environ Microbiol 70:4720–4726
Daane L, Häggblom M (1999) Earthworm egg capsules as vectors for the environmental introduction of biodegradative bacteria. Appl Environ Microbiol 65:2376–2381
Daniel R (2004) The soil metagenome—a rich resource for the discovery of novel natural products. Curr Opin Biotechnol 15:199–204
Dean-Ross D, Moody J, Cerniglia CE (2002) Utilization of mixtures of polycyclic aromatic hydrocarbons by bacteria isolated from contaminated sediment. FEMS Microbiol Ecol 41:1–7
Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, Huffnagle GB (2014) Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol 52:3605–3613
Dong X et al (2019) Metabolic potential of uncultured bacteria and archaea associated with petroleum seepage in deep-sea sediments. Nat Commun 10:1816
Duarte M, Nielsen A, Camarinha-Silva A, Vilchez-Vargas R, Bruls T, Wos-Oxley ML (2017) Functional soil metagenomics: elucidation of polycyclic aromatic hydrocarbon degradation potential following 12 years of in situ bioremediation. Environ Microbiol 19:2992–3011
Dybas MJ et al (2002) Development, operation, and long-term performance of a full-scale biocurtain utilizing bioaugmentation. Environ Sci Technol 36:3635–3644
El-Bestawy E, Sabir J, Mansy A, Zabermawi N (2014) Comparison among the efficiency of different bioremediation technologies of Atrazine-contaminated soils. J Bioremed Biodeg 5:237
Fuentes MS, Benimeli CS, Cuozzo SA, Saez JM, Amoroso MJ (2010) Microorganisms capable to degrade organochlorine pesticides. Curr Res Technol Educ Top Appl Microbiol Microb Biotechnol 2(2):1255–1264
Gangola S, Joshi S, Kumar S, Pandey SC (2019) Comparative analysis of fungal and bacterial enzymes in biodegradation of xenobiotic compounds. Smart bioremediation technologies: microbial enzymes. Academic Press, Cambridge, MA, pp 169–189
Chapter Google Scholar
Garcia-Junco M, Gomez-Lahoz C, Niqui-Arroyo J-L, Ortega-Calvo J-J (2003) Biosurfactant-and biodegradation-enhanced partitioning of polycyclic aromatic hydrocarbons from nonaqueous-phase liquids. Environ Sci Technol 37:2988–2996
Gordillo F, Chavez FP, Jerez CA (2007) Motility and chemotaxis of Pseudomonas sp. B4 towards polychlorobiphenyls and chlorobenzoates. FEMS Microbiol Ecol 60:322–328
Goux S, Shapir N, El Fantroussi S, Lelong S, Agathos SN, Pussemier L (2003) Long-term maintenance of rapid atrazine degradation in soils inoculated with atrazine degraders. Water Air Soil Pollut Focus 3:131–142
Guang-Guo Y (2018) Remediation and mitigation strategies. Integrated analytical approaches for pesticide management. Elsevier, Amsterdam
Gupta G, Chandra A, Varjani SJ, Banerjee C, Kumar V (2018) Role of biosurfactants in enhancing the microbial degradation of pyrene. In: Bioremediation: applications for environmental protection and management. Springer, Singapore
Gupta G, Kumar V, Pal AK (2019) Microbial degradation of high molecular weight polycyclic aromatic hydrocarbons with emphasis on pyrene. Polycycl Aromat Compd 39:124–138
Hall J, Soole K, Bentham R (2011) Hydrocarbon phytoremediation in the family Fabacea—a review. Int J Phytoremediation 13:317–332
Harwood CS, Gibson J (1997) Shedding light on anaerobic benzene ring degradation: a process unique to prokaryotes? J Bacteriol 179:301–309
Holmes DE, O’Neil RA, Chavan MA, N’Guessan LA, Vrionis HA, Perpetua LA (2009) Transcriptome of Geobacter uraniireducens growing in uranium-contaminated subsurface sediments. ISME J 3:216–230
http://dx.doi.org/10.4172/2155-6199.1000248
https://www.mdeq.ms.gov/wp-content/uploads/2017/06/Bioremediation
Jaiswal S, Singh DK, Shukla P (2019) Gene editing and systems biology tools for pesticide bioremediation: a review. Front Microbiol 10:87
Jennings LK, Chartrand MMG, Lacrampe-Couloume G, Lollar BS, Spain JC, Gossett JM (2009) Proteomic and transcriptomic analyses reveal genes upregulated by cis -dichloroethene in Polaromonas sp. strain JS666. Appl Environ Microbiol 75:3733–3744
Jitnuyanont P, Sayavedra-Soto LA, Semprini L (2001) Bioaugmentation of butane-utilizing microorganisms to promote cometabolism of 1,1,1-trichloroethane in groundwater microcosms. Biodegradation 12:11–22
Karami A, Shamsuddin ZH (2010) Phytoremediation of heavy metals with several efficiency enhancer methods. Afr J Biotechnol 9:3689–3698
CAS Google Scholar
Kariyama R, Kumon H (2003) Biofilm infections. Nihon rinsho Jpn J Clin Med 61:266
Keum YS, Seo JS, Li QX, Kim JH (2008) Comparative metabolomic analysis of Sinorhizobium sp. C4 during the degradation of phenanthrene. Appl Microbiol Biotechnol 80:863–872
Kim S-J, Kweon O, Jones RC, Freeman JP, Edmondson RD, Cerniglia CE (2007) Complete and integrated pyrene degradation pathway in Mycobacterium vanbaalenii PYR-1 based on systems biology. J Bacteriol 189:464–472
Kumar A, Bisht B, Joshi V, Dhewa T (2011) Review on bioremediation of polluted environment: a management tool. Int J Environ Sci 1:1079
Lacerda CM, Reardon KF (2009) Environmental proteomics: applications of proteome profiling in environmental microbiology and biotechnology. Brief Funct Genom Proteom 8:75–87
Lambert JM, Yang T, Thomson NR, Barker JF (2009) Pulsed biosparging of a residual fuel source emplaced at CFB borden. Int J Soil Sediment Water 2:6
Lange C et al (2007) Genome-wide analysis of growth phase-dependent translational and transcriptional regulation in halophilic archaea. BMC Genom 8:415
Law AM, Aitken MD (2003) Bacterial chemotaxis to naphthalene desorbing from a nonaqueous liquid. Appl Environ Microbiol 69:5968–5973
Lima D et al (2009) Evaluating a bioremediation tool for atrazine contaminated soils in open soil microcosms: the effectiveness of bioaugmentation and biostimulation approaches. Chemosphere 74:187–192
Luo Q, Zhang X, Wang H, Qian Y (2005) The use of non-uniform electrokinetics to enhance in situ bioremediation of phenol-contaminated soil. J Hazard Mater 121:187–194
MacNaughton SJ, Stephen JR, Venosa AD, Davis GA, Chang YJ, White DC (1999) Microbial population changes during bioremediation of an experimental oil spill. Appl Environ Microbiol 65:3566–3574
Major DW et al (2002) Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ Sci Technol 36:5106–5116
Malik A (2006) Bioremediation. In: Environmental Microbiology. national science digital library, (xth five year plan network project of NISCAIR (CSIR), UGC, MHRD, New Delhi)
Malla MA, Dubey A, Yadav S, Kumar A, Hashem A, Abd Allah EF (2018) Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front Microbiol 9:1132
Martinez A, Kolvek SJ, Yip CL, Hopke J, Brown KA, MacNeil IA, Osburne MS (2004) Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol 70:2452–2463
Meckenstock RU, Morasch B, Warthmann R, Schink B, Annweiler E, Michaelis W, Richnow HH (1999) 13C/12C isotope fractionation of aromatic hydrocarbons during microbial degradation. Environ Microbiol 1:409–414
Mesjasz-Przybylowicz J et al (2004) Uptake of cadmium, lead, nickel and zinc from soil and water solutions by the nickel hyperaccumulator Berkheya coddii . Acta Biol Cracov Bot 46:75–85
Molin S, Klemm P, Poulsen L, Biehl H, Gerdes K, Andersson P (1987) Conditional suicide system for containment of bacteria and plasmids. Nat Biotechnol 5:1315
Naik M, Duraphe M (2012) Review paper on–parameters affecting bioremediation. Int J Life Sci Pharma Res 2:L77–L80
Niu H, Wang J, Zhuang W, Liu D, Chen Y, Zhu C (2018) Comparative transcriptomic and proteomic analysis of Arthrobacter sp. CGMCC 3584 responding to dissolved oxygen for cAMP production. Sci Rep 8:1–13
Odukkathil G, Vasudevan N (2013) Enhanced biodegradation of endosulfan and its major metabolite endosulfate by a biosurfactant producing bacterium. J Environ Sci Health Part B 48:462–469
Olawale A, Akintobi O (2011) Biodegradation of glyphosate pesticide by bacteria isolated from agricultural soil Report and Opinion 3:124–128
Olson MS, Ford RM, Smith JA, Fernandez EJ (2004) Quantification of bacterial chemotaxis in porous media using magnetic resonance imaging. Environ Sci Technol 38:3864–3870
Pande V, Pandey SC, Joshi T, Sati D, Gangola S, Kumar S, Samant M (2019) Biodegradation of toxic dyes: a comparative study of enzyme action in a microbial system. In: Smart bioremediation technologies: microbial enzymes. pp 255
Pandey SC, Pande V, Sati D, Gangola S, Kumar S, Pandey A, Samant M (2019) Microbial keratinase: a tool for bioremediation of feather waste. In: Smart bioremediation technologies: microbial enzyme. pp 217
Pandey SC, Pandey A, Joshi T, Pande V, Sati D, Samant M (2019) Microbiological monitoring in the biodegradation of food waste. in: global initiatives for waste reduction and cutting food loss. In: IGI Global. pp 116–140
Petersen J (2011) Phylogeny and compatibility: plasmid classification in the genomics era. Arch Microbiol 193:313–321
Prescott LM, Harley JP, Klein DA (2002) Microbiology, 5th edn. McGrawHill, New York
Roane TM, Josephson KL, Pepper IL (2001) Dual-bioaugmentation strategy to enhance remediation of cocontaminated soil. Appl Environ Microbiol 67:3208–3215
Rondon MR et al (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66:2541–2547
Roy M, Giri AK, Dutta S, Mukherjee P (2015) Integrated phytobial remediation for sustainable management of arsenic in soil and water. Environ Int 75:180–198
Samant M, Pandey SC, Pandey A (2018) Impact of hazardous waste material on environment and their management strategies. In: Microbial biotechnology in environmental monitoring and cleanup. pp 175–192
Sardrood BP, Goltapeh EM, Varma A (2013) An introduction to bioremediation. Fungi as bioremediators. Springer, Berlin, Heidelberg, pp 3–27
Schloss PD, Handelsman J (2003) Biotechnological prospects from metagenomics. Curr Opin Biotechnol 14:303–310
Shinde S (2013) Bioremediation. Overview Recent Res Sci Technol 5:67–72
Singer A, Gilbert E, Luepromchai E, Crowley D (2000) Bioremediation of polychlorinated biphenyl-contaminated soil using carvone and surfactant-grown bacteria. Appl Microbiol Biotechnol 54:838–843
Garima T, Singh, SP (2016) Application of bioremediation on solid waste management: a review. Solid Waste Manag Policy Plan Sustain Soc 143
Singh D, Fulekar MH (2010) Biodegradation of petroleum hydrocarbons by Pseudomonas putida strain MHF 7109 CLEAN–soil. Air Water 38:781–786
Singh BK, Walker A, Morgan JA, Wright DJ (2004a) Biodegradation of chlorpyrifos by enterobacter strain B-14 and its use in bioremediation of contaminated soils. Appl Environ Microbiol 70:4855–4863
Singh P, Suri CR, Cameotra SS (2004b) Isolation of a member of Acinetobacter species involved in atrazine degradation. Biochem Biophys Res Commun 317:697–702
Smith AE, Hristova K, Wood I, Mackay DM, Lory E, Lorenzana D, Scow KM (2005) Comparison of biostimulation versus bioaugmentation with bacterial strain PM1 for treatment of groundwater contaminated with methyl tertiary butyl ether (MTBE). Environ Health Perspect 113:317–322
Streger SH, Vainberg S, Dong H, Hatzinger PB (2002) Enhancing transport of hydrogenophaga flava ENV735 for bioaugmentation of aquifers contaminated with methyl tert-butyl ether. Appl Environ Microbiol 68:5571–5579
Tang YJ, Martin HG, Dehal PS, Deutschbauer A, Llora X, Meadows A (2009) Metabolic flux analysis of Shewanella spp. reveals evolutionary robustness in central carbon metabolism. Biotechnol Bioeng 102:1161–1169
Tang J, Wang R, Niu X, Zhou Q (2010) Enhancement of soil petroleum remediation by using a combination of ryegrass ( Lolium perenne ) and different microorganisms. Soil Tillage Res 110:87–93
Techtmann SM, Hazen TC (2016) Metagenomic applications in environmental monitoring and bioremediation. J Ind Microbiol Biotechnol 43:1345–1354
Torres B, Jaenecke S, Timmis KN, García JL, Díaz E (2003) A dual lethal system to enhance containment of recombinant micro-organisms. Microbiology 149:3595–3601
Torsvik V, Ovreas L (2002) Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5:240–245
Tripathi M, Singh D, Vikram S, Singh V, Kumar S (2018) Metagenomic approach towards bioprospection of novel biomolecule(s) and environmental bioremediation. Annu Res Rev Biol 22:1–12
Urgun-Demirtas M, Stark B, Pagilla K (2006) Use of genetically engineered microorganisms (GEMs) for the bioremediation of contaminants. Crit Rev Biotechnol 26:145–164
Van Deuren J, Lloyd T, Chhetry S, Raycharn L, Peck J (2002) Remediation technologies screening matrix and reference guide, vol 4. Federal Remediation Technologies Roundtable
Venter JC et al (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, Jansson JK (2009) Shotgun metaproteomics of the human distal gut microbiota. ISME J 3:179
Voget S, Leggewie C, Uesbeck A, Raasch C, Jaeger KE, Streit WR (2003) Prospecting for novel biocatalysts in a soil metagenome. Appl Environ Microbiol 69:6235–6242
Wenderoth D, Rosenbrock P, Abraham W-R, Pieper D, Höfle M (2003) Bacterial community dynamics during biostimulation and bioaugmentation experiments aiming at chlorobenzene degradation in groundwater. Microb Ecol 46:161–176
Yousaf S, Ripka K, Reichenauer T, Andria V, Afzal M, Sessitsch A (2010) Hydrocarbon degradation and plant colonization by selected bacterial strains isolated from Italian ryegrass and birdsfoot trefoil. J Appl Microbiol 109:1389–1401
Zhao B, Yeo CC, Poh CL (2005) Proteome investigation of the global regulatory role of s54 in response to gentisate induction in Pseudomonas alcaligenes NCIMB 9867. Proteomic 5:1868–1876
Zhao X, Hardin IR, Hwang HM (2006) Biodegradation of a model azo disperse dye by the white rot fungus Pleurotus ostreatus . Int Biodeterior Biodegrad 57:1–6
Zhou J et al (2003) Bacterial phylogenetic diversity and a novel candidate division of two humid region, sandy surface soils. Soil Biol Biochem 35:915–924
Download references
Acknowledgements
Authors are thankful to the Department of Zoology, Kumaun University, SSJ Campus, Almora (Uttarakhand), India and for providing facility and space for this research work.
Author information
Veni Pande and Satish Chandra Pandey contributed equally to this study.
Authors and Affiliations
Cell and Molecular Biology Laboratory, Department of Zoology, Kumaun University, SSJ Campus, Almora, Uttarakhand, 263601, India
Veni Pande, Satish Chandra Pandey, Diksha Sati & Mukesh Samant
Department of Biotechnology, Kumaun University, Bhimtal Campus, Bhimtal, Nainital, 263136, Uttarakhand, India
Veni Pande, Satish Chandra Pandey & Veena Pande
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Mukesh Samant .
Ethics declarations
Conflict of interest.
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Reprints and permissions
About this article
Pande, V., Pandey, S.C., Sati, D. et al. Bioremediation: an emerging effective approach towards environment restoration. Environmental Sustainability 3 , 91–103 (2020). https://doi.org/10.1007/s42398-020-00099-w
Download citation
Received : 05 May 2019
Revised : 02 February 2020
Accepted : 04 February 2020
Published : 28 February 2020
Issue Date : March 2020
DOI : https://doi.org/10.1007/s42398-020-00099-w
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Bioremediation
- Biostimulation
- Bioaugmentation
- Find a journal
- Publish with us
- Track your research
An official website of the United States government
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock Locked padlock icon ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
- Publications
- Account settings
- Advanced Search
- Journal List

Recent Advances in Enzymes for the Bioremediation of Pollutants
Seyyed mojtaba mousavi, seyyed alireza hashemi, seyed mohammad iman moezzi, navid ravan, ahmad gholami, chin wei lai, wei-hung chiang, navid omidifar, khadije yousefi, gity behbudi.
- Author information
- Article notes
- Copyright and License information
Academic Editor: Néstor Gutiérrez-Méndez
Corresponding author.
Received 2021 Jan 26; Revised 2021 May 5; Accepted 2021 Jun 9; Collection date 2021.
This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Nowadays, pollution of the environment is a huge problem for humans and other organisms' health. Conventional methods of pollutant removal like membrane filtration or ion exchange are not efficient enough to lower the number of pollutants to standard levels. Biological methods, because of their higher efficiency and biocompatibility, are preferred for the remediation of pollutants. These cost-effective and environment-friendly methods of reducing pollutants are called bioremediation. In bioremediation methods, enzymes play the most crucial role. Enzymes can remedy different types of organic and inorganic pollutants, including PAHs, azo dyes, polymers, organocyanides, lead, chromium, and mercury. Different enzymes isolated from various species have been used for the bioremediation of pollutants. Discovering new enzymes and new subtypes with specific physicochemical characteristics would be a promising way to find more efficient and cost-effective tools for the remediation of pollutants.

1. Introduction
The widespread use of chemicals in industries and militaries, inadequate waste disposal, and accidental leakage cause contamination of soil, water, and air. For instance, there are 34,000 contaminated sites just in Europe that need to be treated. These pollutants are hazardous for humans, other living beings, and even the biogeochemical cycle. Pollutants' stability, low solubility, and resistance to various physical, chemical, and biological degradation pathways are the main reasons for their toxicity [ 1 ].
Different physical and chemical methods for cleaning up pollutants have been used, such as oxidizing agents, electrochemical treatments, adsorption of pollutants, ion exchange, and membrane filtration [ 2 ]. Despite the adequacy of traditional methods for the high concentration of pollutants, they were not enough for lowering the amount of contamination to regulatory limits [ 3 ]. Various disadvantages of traditional methods for cleaning up pollutants include high cost, nonspecificity, and probable secondary contamination production; therefore, ecofriendly and biological methods, called bioremediation, gained interest [ 4 ].
Bioremediation is defined as processes and products that are cost-effective and practical to minimize pollutants in the source and diminish danger to the environment and human health [ 5 ]. Its main ways of degrading and detoxifying pollutants are through intracellular accumulation or enzymatic transformation [ 4 ]. Pollutant properties (i.e., chemical structure, hydrophobicity, and polarity), environmental conditions (i.e., temperature, pH, and redox condition), and soil features (i.e., aggregation, thickness, dissolved organic matter, and pollutants aging) affect biological degradation and contaminants availability [ 6 ].
Enzymes are the most efficient bioremediation tools and progress all chemical changes on pollutants. Enzymes' specificity is usually broad enough to act on different molecules with similar structures. Moreover, it is possible to engineer the enzymes for enhancing their stability and efficiency for special conditions or particular substrates [ 7 , 8 ]. Omics technologies have a significant role in these developments [ 2 ].
Using enzymes in bioremediation could be either individually that the isolated enzyme used and added to the contaminated area or as a whole cell, e.g., bacteria, fungi, or algae. In a second way, continuous aeration, inoculation, and nutrition are necessary. Besides, environmental conditions should be prepared for microorganisms living, even though there might still be some toxic compounds in the environment that inhibit microorganisms' activity [ 1 , 9 ]. The use of individual enzymes has some advantages in comparison with microbial whole cell including greater specificity, more straightforward handling and storage, standardizable activity, more mobility as a result of smaller size, being active in the presence of high concentrations of toxic compounds, and biodegradability that inhibits persistence and recalcitrance [ 1 , 10 , 11 ]. This approach is much more efficient for extracellular enzymes and cofactor-independent enzymes [ 12 , 13 ].
Enzyme production in the natural environment is low, while it is possible to increase the produced enzyme under controlled conditions. On the other hand, recombinant DNA technology and gene engineering provide many opportunities to produce more efficient and more enzymes [ 14 ]. Moreover, nanotechnology offers some tools to increase enzymes' stability by decreasing sensitivity to mechanical stress, preserving the third structure of enzymes, and protecting them against proteases [ 9 ].
Enzymatic bioremediation could be in situ or ex situ . In in situ methods with the least disturbance in the environment, the free or immobilized enzyme (adsorbed enzymes on mineral supports that minimize the loss of enzymatic activity) is added to the soil. This approach is less expensive because of no need for excavation and transportation of soil. Ex situ methods are feasible for highly contaminated soils with toxic pollutants or when fast action is essential. During this procedure, soil was excavated and treated in different bioreactors in the best condition for enzymes' activity [ 1 ] ( Figure 1 ).
A representative scheme of different methods of soil remediation. (a) Conventional in situ remediation. (b) Using a single-phase bioreactor for solvent extraction. (c) Using a two-phase bioreactor for solvent extraction. Adapted from [ 1 ].
Different enzymes like mono- or dioxygenases, halogenases, peroxidases, phosphotriesterases, hydrolases, transferases, and oxidoreductases from various species of bacteria, fungi, algae, and plants have been used for the bioremediation of pollutants [ 10 , 15 ]. We try to review the most essential enzymes for the bioremediation of pollutants and insight into their mechanism of action.
2. Enzymes for Organic Substrates
Large amounts of organic pollutants, including herbicides, pesticides, dyes, drugs, and plastics, pollute the air, soil, and water every year. Polymers, aromatic molecules, polycyclic aromatic hydrocarbons (PAHs), chlorinated hydrocarbons, steroids, and organocyanides are the most organic compounds that need to be cleaned up worldwide. Their stable structure is the main reason for their toxicity.
2.1. Hydrolases (EC 3)
Esterases, nitrilases, aminohydrolases, lipase, cutinase, and organophosphorus hydrolase are among the hydrolase enzymes used in the bioremediation of different chemicals such as herbicides, pesticides, organophosphorus compounds, nitrile compounds, and polymers [ 1 , 2 ]. We would review some of them shortly as follows.
2.2. Esterases (3.1)
Esterases catalyze the cleavage of ester bonds in different chemicals like organophosphorus herbicides and pesticides, diethyl glycol adipate, polyurethanes, and aromatic and aliphatic polyesters. Escherichia coli and Pichia pastoris are two bacteria that express and colonize the thermostable kind of enzymes. Moreover, a subgroup of esterases found in E. coli is active in a cold environment and can act on phthalate esters [ 2 ].
It is worth noting that the product of esterase reaction with organophosphorus compounds, 3,5,6-trichloro-2-pyridinol (TCP), is metabolized later to less toxic chemicals by aminohydrolase (EC 3.5) [ 2 ].
2.3. Nitrilases (EC 3.5.5.1)
Triple bonds between carbon and nitrogen (nitrile group) of herbicides, polymers, and plastics are hydrolyzed stereo-, regio-, or chemoselectively by nitrilases to carboxylic acid and ammonia. Many species can express these enzymes, including Streptomyces sp., Fusarium solani, Rhodococcus rhodochrous, Aspergillus niger, Bacillus pallidus, and Pseudomonas fluorescens . Moreover, an evolution approach on Alcaligenes faecalis tends to isolate a nitrilase that was active in the broader range of pH. Besides, P. fluorescens nitrilase's gene expressed in E. coli is probably the most hopeful nitrilase [ 16 , 17 ]. Cyanide dihydratase (EC 3.5.5) is one of the nitrilases and degrade cyanide into formate and ammonia. Pseudomonas stutzeri and Bacillus pumilus are two species that express this enzyme. Furthermore, fungal cyanide hydratase (EC 4.2.1.66), isolated from Fusarium lateritium, Neurospora crassa , and Gloeocercospora sorghi , and some other species, is another cyanide-degrading enzyme that metabolizes it to formamide [ 18 ]. These enzymes are promising for the bioremediation of wastewaters from coal coking and metal-plating baths [ 17 ].
2.4. Organophosphorus Hydrolase (EC 3.1.8.2)
Organophosphate compounds were developed and used as pesticides and in warfare and even as a drug since 1937. They are neurotoxic, and after a while, they were more than that soil microbiota could remedy all of them. Organophosphorus hydrolase (also known as phosphotriesterase) is one of the enzymes that can serve for organophosphorus compounds bioremediation. It is mostly isolated from Pseudomonas diminuta , although its fungal form is expressed in Aspergillus niger and Penicillium lilacinum . It can act on P-S, P-O, and P-F bonds. This enzyme has Zn 2+ as a cofactor in its native form, while assays showed that substitution of Co 2+ provides the most potent activity against paraoxon [ 19 ]. This enzyme has the fastest catalytic rate and is the most promising enzyme for engineering activity against organophosphates [ 20 ].
2.4.1. Peroxidases
(1) Ligninolytic Peroxidases . Ligninolytic enzymes are a family of enzymes with broad applications in bioremediation. This group of enzymes produced by white-rot fungi (WRF) is in the condition of nutrient limitation known as “ligninolytic.” Also, lignocellulosic materials can be an inducer for the production of these enzymes [ 21 ]. Due to the high nonspecificity and high nonstereoselectivity of these enzymes, they can degrade a wide range of recalcitrant compounds [ 22 ]. They degrade chemicals by pseudo-first-order kinetic via a free-radical-based chain reaction using H 2 O 2 and molecular oxygen [ 21 – 24 ].
Ligninolytic enzymes can be categorized into four main enzymes, including laccase (LAC), lignin peroxidase (LiP), manganese peroxidase (MnP), and versatile peroxidase (VP).
(2) Laccase . For oxidizing phenolic compounds, PAHs, dyes, and pesticides benzenediol: oxygen oxidoreductase, known as laccase, is a suitable enzyme. As an oxidase, laccase substrates go through one of the following pathways: (1) cleavage of aromatic rings, (2) polymerization, and (3) degradation of covalent bonds between monomers. Four atoms of copper are the principal part of the reaction, and oxygen is the last electron receptor [ 2 , 25 ]. The mechanism of the reaction is shown in Figure 2 .
General reaction mechanism of bacterial laccases. Adapted from [ 26 ].
Laccase is first discovered in different fungi species like Panus conchatus and Polyporus sp. Later on, laccase was found in Azospirillum lipoferum, as the first bacteria species. Laccase is produced in different Gram-positive bacteria, including Bacillus , Geobacillus, Aquisalibacillus, Lysinibacillus, Staphylococcus , and Streptomyces . Many bacteria produce laccase extracellularly, while some others are unable to secrete the enzyme. Bacterial laccase is more resistant to extreme temperature and pH conditions [ 1 , 25 ].
There are two kinds of laccase, white and blue. The main difference between these is that blue laccase is dependent on a “mediator” for the degradation of nonphenolic substrates. “Mediator” is an intermediator that laccase oxidizes and turns into oxidized radicals that react with high redox potential or bulky substrates. ABTS (2,20-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid)) and N-heterocycles with N-OH such as violuric acid, N-hydroxybenzotriazole, and N-hydroxy-N-phenylacetamide have been used as effective mediators [ 25 ].
Every year, approximately 7 × 10 4 − 1 × 10 7 tons of dyes penetrate the environment [ 25 ]. Laccase is used for dyes remediation. As an example, a Bacillus licheniformis LS40-derived laccase can decolorize azo, indigo, and anthraquinone dyes by 80% within one hour in the presence of acetosyringone as a mediator [ 27 ].
PAHs are xenobiotic pollutants because of their low solubility and degradation rate. Laccase can convert PAHs to their less toxic quinine form and CO 2 . There are some examples in Table 1 . Notably, laccase can degrade some drugs such as diclofenac and mefenamic acid in acidic pH [ 25 ].
Examples of laccase's pollutant bioremediation.
(3) Lignin Peroxidase . Lignin peroxidases (LiPs) are a group of heme-containing monomeric enzymes. Their weight ranges between 38 and 43 kDa [ 30 , 31 ] with iron in the ferric state [ 21 , 32 ]. LiPs with their high redox potential [ 32 ] are capable of breaking alpha and beta carbon bonds, catalyzing the degradation of phenolic and nonphenolic compounds, demethylation, and opening aromatic ring of dyes [ 33 ]. LiPs have a high redox potential for oxidizing nonphenolic structures [ 31 ].
LiP activity increases in the presence of H 2 O 2 as an electron acceptor. However, high concentrations of H 2 O 2 could damage the LiPs [ 32 ]. In the first step of the reaction, Fe 3+ binds to H 2 O 2 and oxo-ferryl intermediate named compound I forms. Then, compound I, by a donation of one electron from the substrate, reduces to compound II, finally; by another electron donation from the substrate, iron in heme returns to its ferric resting state, and the enzyme renews to its initial form [ 31 , 34 ]. In this three-step reaction, the reduction of compound II is the rate-limiting step ( Figure 3 ) [ 36 ]. Due to this slow reduction rate, compound I is available for reaction with H 2 O 2 and the formation of a complex between LiP and superoxide (compound III) inactive enzyme [ 36 ].
Lignin peroxides catalytic reaction. Adapted from [ 35 ].
Veratryl alcohol is a secondary metabolite that can play essential roles in this oxidizing reaction. Veratryl alcohol can be the mediator in the electron transfer reaction; it can play a role in the catalytic cycle of LiP by an oxidizing terminal substrate. Vertaryl alcohol can also prevent the formation of compound III and, if compound III is established, reduce it to its native form [ 36 ].
Many WRFs produce LiPs such as Phanerochaete chrysosporium , Trametes versicolor , Bjerkandera adusta , Phlebia radiate , and Ganoderma lucidum [ 22 , 31 ].
Many technologies have been applied to enhance activity and increase catalytic characteristics of LiPs, such as LiP entrapment in calcium beads [ 37 ].
(4) Manganese Peroxidase . Manganese peroxidases (MnPs) are heme-containing glycol proteins with weight ranging from 32 to 62.5 kDa [ 38 ]. Like other ligninolytic peroxidases, MnP uses H 2 O 2 . By using H 2 O 2 , MnP can oxidase Mn 2+ to Mn 3+ . The first step of the reaction is binding an oxygen atom of H 2 O 2 to Fe 3+ of heme. Then, by two-electron transfer from Fe 3+ to peroxide Fe 4+ oxo-porphyrin, compound I radical forms. Then, compound I binds to monochelated Mn 2+ and Mn 3+ and compound II forms. Finally, by oxidizing another Mn 2+ to Mn 3+ , compound II reduces and the enzyme with Fe 3+ reforms ( Figure 4 ) [ 32 , 36 ].
Manganese peroxidase catalytic reaction. Adapted from [ 35 ].
Aliphatic organic acids such as lactate and oxalate can induce Mn 2+ oxidation rate, and Mn 3+ -acid chelates have a higher redox potential. MnP activity increases in the presence of glutathione and unsaturated fatty acids, such as tween 80. Many techniques have been utilized to immobilize and enhance the efficacy of bioremediation with MnP, such as making calcium alginate beads and carbon nanotubes [ 39 – 41 ].
MnP can remediate PAHs and nitroaromatic compounds [ 36 , 42 ], azo dyes [ 43 ], and endocrine-disrupting chemicals such as bisphenol A and alkylphenols [ 44 , 45 ]; moreover, with the contribution of mediators such as lipid and thiyl radicals, MnP is capable of oxidizing nonphenolic structures [ 34 ].
Many species of fungi are able to produce MnP, such as Phanerochaete chrysosporium , Trametes versicolor , Irpex lacteus , Dichomitus squalens , and Ganoderma lucidum [ 45 , 46 ].
(5) Versatile Peroxidase . Versatile peroxidase (VP) is a heme-containing ligninolytic enzyme considered as a hybrid between LiP and MnP. VP has two active sites; therefore, it can oxidize both Mn 2+ and veratryl alcohol by a similar mechanism to MnP and LiP, respectively [ 32 , 47 ].
VP can oxidize both low and high redox potential compounds, polycyclic aromatic hydrocarbons, azo dyes, high molecular weight aromatics, and both phenolic and nonphenolic compounds and environmental pollutants [ 32 , 47 , 48 ].
VP production is less common in WRFs than MnP and LiP, but it can be found in some species such as Pleurotus spp. and Bjerkandera spp. [ 32 ].
2.4.2. Horseradish Peroxidase
Horseradish peroxidase (HRP) is an enzyme traditionally extracted and isolated from the root of horseradish ( Armoracia rusticana ). The most abundant isoenzyme found in the root of horseradish is C isoenzyme (HRPC). HPRC is 44 kDa heme-containing glycopeptide with 308 amino acids, an iron atom in the ferric state in protoporphyrin IX, and two calcium atoms in the central zone [ 49 – 51 ]. HRP catalyzes oxidative reaction using H 2 O 2 . In the presence of H 2 O 2 , the intermediate compound formed via two-electron oxidation. Then by an oxidable substrate, compound I reduces to compound II. Radical formation occurs via these reactions, and finally, the initial enzyme can be renewed by the reaction of compound II with another substrate molecule. In comparison with LiP compound, I and II are more electronegative in HRP ( Figure 5 ) [ 52 ].
Horseradish peroxidase catalytic cycle. Adapted from [ 52 ].
HRP is applicable for removing and remediating phenols, substrate phenols, and alkylphenols, aromatic amines [ 53 , 54 ], azo dyes [ 55 , 56 ], endocrine-disrupting compounds [ 54 ], and many other environmental pollutants.
Many techniques have been utilized to immobilize and enhance the efficacy of enzyme by nanotechnology [ 57 – 59 ]. Using horseradish root is a standard method; using fertile soil for horseradish cultivation to feed the population has raised concern in recent years [ 60 ]. To solve this problem and enhance the efficacy of the enzyme, many biotechnological methods have been experienced, such as recombinant production of HRP in E. coli , yeast, plants, and insect systems [ 61 ].
2.5. Cytochrome p450 Monooxygenase (EC 1.14.14.1)
Cytochrome p450 monooxygenases (CYP) are a family of heme-containing enzymes that catalyze different reactions such as N-hydroxylation, N-dealkylation, O-dealkylation, oxidative dehalogenation, and hydroxylation of C-H bonds. CYP derives essential electrons for reactions from NADPH-cytochrome p450 reductase, and the latter enzyme derives electrons from atmospheric oxygen. So, the presence of a reducing agent like NAD (P) H or FAD is necessary [ 62 ]. The reaction cycle of CYP450 is shown in Figure 6 .
Cytochrome p450 reaction cycle. RH: substrate; ROH: product. Adapted from [ 63 ].
CYPs are versatile enzymes expressed in various species of bacteria, fungi, plants, and animals. About 7000 different CYPs have been discovered till now. Saccharomyces, Streptomyces, Basidiomycete, Dehalococcoides, Rhodococcus, Bacillus, Escherichia, and Salmonella are among the genera that their CYPs are used for bioremediation [ 64 , 65 ].
While bacterial CYPs are attractive because of their solubility, easy and low-cost production, and self-efficiency (their electron transfer reductases, e.g., FMN, FAD, and p450 monooxygenase, are on a single peptide), mammalian CYPs are membrane-bounded, dependent on a redox partner (e.g., NADPH) and have expansive applications [ 65 ]. Bacterial and eukaryotic CYPs can oxidize aliphatic hydrocarbons with 5–16 and 10–16 carbon lengths, respectively [ 66 ]. Notably, eukaryotic CYPs need modification at N-terminal, but prokaryotic ones are active in the native form [ 64 ].
Dioxins, PCBs (polychlorinated biphenyls), PCDDs (polychlorinated dibenzo-p-dioxins), PCDFs (polychlorinated dibenzofurans), PAHs, aliphatic hydrocarbons, and even Cr (VI) are pollutants that can be degraded and bioremedied by CYPs [ 1 , 14 , 65 , 67 ]. In Table 2 , the list of different CYPs and their substrates are shown. Immobilizing CYPs can improve their activity even to 10-folds higher than free enzyme. Besides, transgenic plants that can produce special CYPs are a way toward herbicide-resistant plants [ 65 ].
Cytochrome p450 subtypes and their substrate for bioremediation.
CYPs are interesting enzymes for bioremediation because of their wide range of substrates and diverse oxidative reactions. Among the limitations of using CYPs are their dependency on expensive cofactors, low stability, and low activity [ 68 ].
3. Enzymes for Inorganic Substrates
In the presence of toxic heavy metals, most of the microorganisms produce metal-binding peptides such as phytochelatins and metallothioneins, which reduce their toxicity via sequestration [ 70 ]. For example, phytochelatin synthase is the enzyme responsible for the production of phytochelatin that, in cooperation with GSH, accumulates heavy metals [ 71 ]. Among the limitations of these metal-binding proteins is their nonselectivity. To solve this problem, many microorganisms developed specific pathways for resistance against heavy metals [ 4 ]. Obviously, enzymes are the most critical part of these pathways; we would review some of these metal-specific enzymes as follows.
3.1. Arsenic
Arsenic is a heavy metal that exists in nature in organic and inorganic forms. The inorganic forms (As 3+ (arsenite) and As 5+ (arsenate)) are toxic and may cause enzyme inactivation, carcinoma, hemolysis, keratosis, gangrene, and neurological and cardiovascular diseases [ 72 , 73 ]. Arsenate and arsenite convert to each other by arsenate reductase and arsenite oxidase through redox reactions. As 3+ is more mobile and toxic. As 5+ is the terminal electron acceptor in the absence of oxygen and reduces to As 3+ [ 63 ]. Ferredoxin or glutathione would be the electron source [ 74 ]. This process enhances the solubility of As and eases leaching from soil [ 73 ]. The final As 3+ is excreted through efflux pumps, ArsB and Acr3 [ 74 ]. Arsenite oxidase converts As 3+ to less toxic As 5+ to be used either for a supplemental energy source or as an electron donor for CO 2 fixation [ 74 ]. The final arsenate is immobile and would be retained by sediments [ 73 ].
The methylated form of arsenic is volatile and would be lost from the soil [ 73 ]. Interestingly, in methanogenic bacteria, As methylation is coupled with methane biosynthesis and can detoxify soil through this mechanism. Coenzyme M is the biocatalyst of this detoxification process [ 63 ].
Many species can remedy As in different ways. The bacterial ones include Acinetobacter sp., Pseudomonas sp., and Sporosarcina ginsengisoli [ 75 ]. E. coli, Bacillus idriensis , and Sphingomonas desiccabilis are engineered species for As bioremediation [ 72 ]. Some fungi, including Rhizobium sp., Rhizopus sp. , Trichoderma sp., Aspergillus flavus , and Penicillium canescens , are As bioremediators too [ 63 , 73 ]. Moreover, some yeasts like Saccharomyces cerevisiae can reduce arsenate by ArsC ( Figure 7 ), a protein that has As reductase activity. Algae genera, like Hydrodictyon, Oedogonium, Rhizoclonium, and even a plant, Pteris vittata from Pteridaceae, have the potential to be used for bioremediation [ 75 ].
A strategy used for arsenic detoxification using E. coli and S. cerevisiae and their enzymes. Adapted from [ 63 ].
Lead was found in a small amount in nature before industrialization. However, now, through gasoline burning, different Pb salts originate in and contaminate water, soil, and air [ 72 ]. Lead toxicity may cause anemia and appetite loss and gastrointestinal, neurological, and reproductive disorders [ 73 ]. Organoleads, especially tetraethyl lead and tetramethyl lead used in gasoline, are toxic forms of lead. They are sensitive to photolysis and volatilization and degrade to dialkyl species. Though, some bacteria can degrade organoleads through bioremediation processes [ 76 ].
Cupriavidus metallidurans can remove Pb 2+ ions with p-type ATPase and produce inorganic phosphate to sequester Pb 2+ in the periplasm [ 76 ]. Staphylococcus epidermidis can biomineralize Pb 2+ by carbonate. Urease enzymes form different carbonate crystalline Pb 2+ . It can be mineralized as oxalate and pyromorphite, too [ 77 ]. Agaricus bisporus, Rhizopus nigricans, Penicillium canescens, Penicillium chrysogenum, Saccharomyces cerevisiae, Aspergillus niger, and Aspergillus terreus are among biotransforming organisms [ 72 , 73 ]. Moreover, it is reported that Arthrobacter and Phaeolus schweinitzii can degrade trimethyl lead cations [ 78 ].
3.3. Mercury
Mercury is a heavy metal that is toxic in both organic and inorganic forms, although the organic form is more toxic. Hg toxicity would cause neurotoxicity, nephrotoxicity, allergies, and inability to speak [ 73 , 79 ]. Hg is a rare element in Earth crust, but it spreads and pollutes soil and water because of different humic activities like gold mining, various measurement tools (barometer, thermometer, manometer, etc.), lamps, mercurial fungicides, paper manufacturing industry, and battery cells [ 72 ]. Its environmental cycle is shown in Figure 8 .
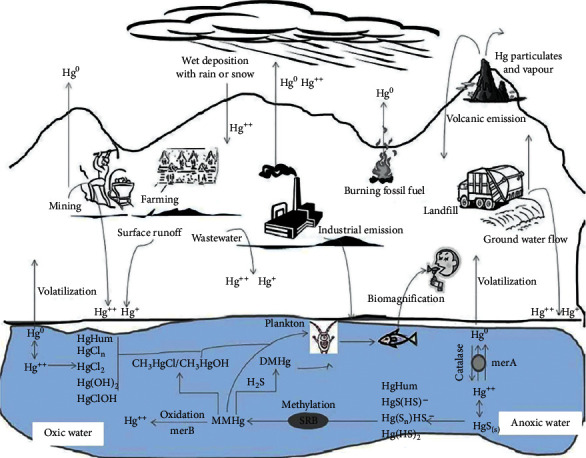
Mercury cycle in the environment. Adapted from [ 80 ].
Mercury exists in three forms: metallic mercury (Hg 0 ), mercurous (Hg +1 ), and mercuric (Hg 2+ ) forms. The most toxic form of Hg is mercuric chloride. Organic mercury can accumulate in living organisms and has an affinity for proteins' sulfhydryl groups. Inorganic mercury has the lowest toxicity because of its low solubility and high vapor pressure. Mercury-resistant bacteria (such as Pseudomonas, Aeromonas, Staphylococcus, Escherichia, Citrobacter, Bacillus, and Rhodococcus ) can reduce toxic organic forms of Hg to less toxic metallic Hg. Mercuric reductase is the main enzyme that reduces Hg. The mer operon is the collection of mercury-resistance genes activated in the presence of an inducible concentration of Hg. Mercuric reductase in cooperation with FAD and NADPH, as electron sources, reduces Hg 2+ to Hg 0 . The final metallic mercury is volatile and spreads to the atmosphere [ 80 , 81 ]. Also, dimethylmercury is volatile and biomethylation can serve as a strategy for Hg bioremediation [ 73 ]. The mer operon-independent volatilization of mercury has been discovered, too, in Shewanella oneidensis [ 82 ].
Another enzyme that plays a role in mercury bioremediation is organomercurial lyase that breaks the carbon-mercury bonds in organo-Hg compounds [ 80 , 81 ].
Various microorganisms such as Rhizopus arrhizus, Penicillium canescens, Geobacter sulfurreducens, Pseudomonas putida, Acinetobacter calcoaceticus, Staphylococcus aureus, and Shigella flexneri can remedy mercury [ 72 , 80 ]. Enterobacter, Pseudomonas , and Bacillus are the most used genera for this purpose [ 83 ].
3.4. Chromium
Cr (VI) is the most toxic heavy metal because of its high oxidative potential causing cell damage and mutagenic, carcinogenic, and teratogenic effects [ 84 ]. The wide use of chromium and its compounds and mining exerts this pollutant to waters and soils. Bioremediation of hexavalent chromium is through reduction to trivalent species. Pseudomonas, Bacillus, Escherichia, Shewanella, Enterobacter , and Thermus are some genera that are resistant to Cr (VI) and can reduce it. The reduction of hexavalent chromium may occur through aerobic or anaerobic pathways [ 14 ]. In the anaerobic process, soluble cytoplasmic enzymes are involved and reduce hexavalent chromium in two steps. In the aerobic reduction of chromium, usually, Cr (VI) is a terminal electron acceptor, while in different species, NADPH, NADH, or formate serves as an electron donor. Chromate reductase, Ni-Fe dehydrogenase, and cytochrome c3 are among the enzymes reported to have hexavalent chromium-reducing activity [ 85 ]. Also, Fe 2+ and S 2- produced in some bacteria can reduce Cr 6+ even faster than chromate-reducing bacteria. The detailed mechanism of chromate resistance in bacteria is shown in Figure 9 .
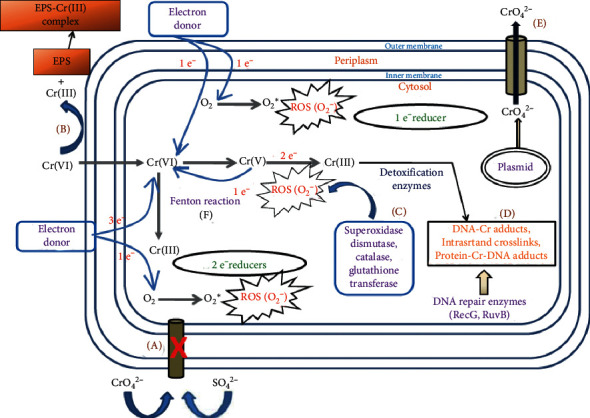
Chromate resistance mechanism in bacteria. (A) Mutation in sulfate uptake transporters. (B) Extracellular reduction of Cr 6+ to Cr 3+ . (C) Intracellular reduction of Cr 6+ to Cr 3+ by chromate reductase. (D) Reducing oxidative stress and activation of repairing systems. (E) Outflowing of chromate from the cytoplasm. (F) Decreasing oxidative stress by activation of ROS scavenging enzyme. Adapted from [ 14 ].
Nitroreductase, iron reductase, flavin reductases, and quinone reductases are bacterial enzymes that reduce Cr 6+ [ 86 , 87 ]. Mammals reduce this pollutant, too, by CYP, aldehyde oxidase, and DT-diaphorase. Some of these bacterial enzymes are extracellular, including nitrate reductases, flavin reductases, and ferrireductases [ 14 ].
4. Conclusion
In this review, we aim to provide an insight into the role of the enzyme in the bioremediation of pollutants. While many physical and chemical methods of treating contaminated soil and water are not efficient enough, bioremediation opens a new way to clean up toxic pollutants. Enzymes as practical tools of living organisms are an ecofriendly and bio-based strategy for bioremediation. Microorganisms exposed to contaminated sites and specific pollutants are fascinating sources for the isolation of active enzymes against those pollutants. Interestingly, we may find some enzymes in completely irrelevant places to pollutant sources. Discovering TCP-degrading enzymes and chlorpyrifos-degrading enzymes in the cow rumen microbiome is an instance for this claim [ 2 ].
Overall, using enzymes for pollutant bioremediation seems to be a cost-effective, efficient, and practical approach. Although there are still many ways to go, further studies and experiments on enzyme activity and mechanism of action and isolating new enzymes would be a promising way to reduce pollutants and make a healthier environment for humans and all other species.
Contributor Information
Ahmad Gholami, Email: [email protected].
Chin Wei Lai, Email: [email protected].
Wei-Hung Chiang, Email: [email protected].
Data Availability
The data used to support this study are available upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
- 1. Eibes G., Arca-Ramos A., Feijoo G., Lema J. M., Moreira M. T. Enzymatic technologies for remediation of hydrophobic organic pollutants in soil. Applied Microbiology and Biotechnology. 2015;99(21):8815–8829. doi: 10.1007/s00253-015-6872-y. [ DOI ] [ PubMed ] [ Google Scholar ]
- 2. Ufarté L., Laville É., Duquesne S., Potocki-Veronese G. Metagenomics for the discovery of pollutant degrading enzymes. Biotechnology Advances. 2015;33(8):1845–1854. doi: 10.1016/j.biotechadv.2015.10.009. [ DOI ] [ PubMed ] [ Google Scholar ]
- 3. Malik A. Metal bioremediation through growing cells. Environment International. 2004;30(2):261–278. doi: 10.1016/j.envint.2003.08.001. [ DOI ] [ PubMed ] [ Google Scholar ]
- 4. Singh S., Kang S. H., Mulchandani A., Chen W. Bioremediation: environmental clean-up through pathway engineering. Current Opinion in Biotechnology. 2008;19(5):437–444. doi: 10.1016/j.copbio.2008.07.012. [ DOI ] [ PubMed ] [ Google Scholar ]
- 5. Kirchhoff M. M. Promoting green engineering through green chemistry. Environmental Science & Technology. 2003;37(23):5349–5353. doi: 10.1021/es0346072. [ DOI ] [ PubMed ] [ Google Scholar ]
- 6. Rao M. A., Scelza R., Acevedo F., Diez M. C., Gianfreda L. Enzymes as useful tools for environmental purposes. Chemosphere. 2014;107:145–162. doi: 10.1016/j.chemosphere.2013.12.059. [ DOI ] [ PubMed ] [ Google Scholar ]
- 7. Theerachat M., Emond S., Cambon E., et al. Engineering and production of laccase from Trametes versicolor in the yeast Yarrowia lipolytica. Bioresource Technology. 2012;125:267–274. doi: 10.1016/j.biortech.2012.07.117. [ DOI ] [ PubMed ] [ Google Scholar ]
- 8. Festa G., Autore F., Fraternali F., Giardina P., Sannia G. Development of new laccases by directed evolution: functional and computational analyses. Proteins: Structure, Function, and Bioinformatics. 2008;72(1):25–34. doi: 10.1002/prot.21889. [ DOI ] [ PubMed ] [ Google Scholar ]
- 9. Rayu S., Karpouzas D. G., Singh B. K. Emerging technologies in bioremediation: constraints and opportunities. Biodegradation. 2012;23(6):917–926. doi: 10.1007/s10532-012-9576-3. [ DOI ] [ PubMed ] [ Google Scholar ]
- 10. Rao M. Role of enzymes in the remediation of polluted environments. Journal of Soil Science and Plant Nutrition. 2010;10(3):333–353. doi: 10.4067/s0718-95162010000100008. [ DOI ] [ Google Scholar ]
- 11. Gianfreda L., Rao M. A. Potential of extra cellular enzymes in remediation of polluted soils: a review. Enzyme and Microbial Technology. 2004;35(4):339–354. doi: 10.1016/j.enzmictec.2004.05.006. [ DOI ] [ Google Scholar ]
- 12. Scott C., Pandey G., Hartley C. J., et al. The enzymatic basis for pesticide bioremediation. Indian Journal of Microbiology. 2008;48(1):65–79. doi: 10.1007/s12088-008-0007-4. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 13. Sutherland T., Horne I., Weir K., et al. Enzymatic bioremediation: from enzyme discovery to applications. Clinical and Experimental Pharmacology and Physiology. 2004;31(11):817–821. doi: 10.1111/j.1440-1681.2004.04088.x. [ DOI ] [ PubMed ] [ Google Scholar ]
- 14. Thatoi H., Das S., Mishra J., Rath B. P., Das N. Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review. Journal of Environmental Management. 2014;146:383–399. doi: 10.1016/j.jenvman.2014.07.014. [ DOI ] [ PubMed ] [ Google Scholar ]
- 15. Pieper D. H., Martins dos Santos V. A., Golyshin P. N. Genomic and mechanistic insights into the biodegradation of organic pollutants. Current Opinion in Biotechnology. 2004;15(3):215–224. doi: 10.1016/j.copbio.2004.03.008. [ DOI ] [ PubMed ] [ Google Scholar ]
- 16. Nigam V. K., Arfi T., Kumar V., Shukla P. Bioengineering of nitrilases towards its use as green catalyst: applications and perspectives. Indian Journal of Microbiology. 2017;57(2):131–138. doi: 10.1007/s12088-017-0645-5. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 17. Martínková L., Rucká L., Nešvera J., Pátek M. Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World Journal of Microbiology and Biotechnology. 2017;33(1):p. 8. doi: 10.1007/s11274-016-2173-6. [ DOI ] [ PubMed ] [ Google Scholar ]
- 18. Park J. M., Trevor Sewell B., Benedik M. J. Cyanide bioremediation: the potential of engineered nitrilases. Applied Microbiology and Biotechnology. 2017;101(8):3029–3042. doi: 10.1007/s00253-017-8204-x. [ DOI ] [ PubMed ] [ Google Scholar ]
- 19. Singh B. K., Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiology Reviews. 2006;30(3):428–471. doi: 10.1111/j.1574-6976.2006.00018.x. [ DOI ] [ PubMed ] [ Google Scholar ]
- 20. Thakur M., Medintz I. L., Walper S. A. Enzymatic bioremediation of organophosphate compounds-progress and remaining challenges. Frontiers in Bioengineering and Biotechnology. 2019;7:p. 289. doi: 10.3389/fbioe.2019.00289. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 21. Barr D. P., Aust S. D. Pollutant degradation by white rot fungi. Reviews of Environmental Contamination and Toxicology. 1994;138:49–72. doi: 10.1007/978-1-4612-2672-7_3. [ DOI ] [ PubMed ] [ Google Scholar ]
- 22. Kaur H., Kapoor S., Kaur G. Application of ligninolytic potentials of a white-rot fungus Ganoderma lucidum for degradation of lindane. Environmental Monitoring and Assessment. 2016;188(10):p. 588. doi: 10.1007/s10661-016-5606-7. [ DOI ] [ PubMed ] [ Google Scholar ]
- 23. Reddy C. A., Mathew Z. British Mycological Society Symposium Series. Amsterdam, Netherlands: Elsevier; 2001. Bioremediation potential of white rot fungi. [ Google Scholar ]
- 24. Pointing S. B. Feasibility of bioremediation by white-rot fungi. Applied Microbiology and Biotechnology. 2001;57(1-2):20–33. doi: 10.1007/s002530100745. [ DOI ] [ PubMed ] [ Google Scholar ]
- 25. Chauhan P. S., Goradia B., Saxena A. Bacterial laccase: recent update on production, properties and industrial applications. 3 Biotech. 2017;7(5):p. 323. doi: 10.1007/s13205-017-0955-7. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 26. Solomon E. I., Augustine A. J., Yoon J. O2 reduction to H2O by the multicopper oxidases. Dalton Transactions. 2008;30(30):3921–3932. doi: 10.1039/b800799c. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 27. Lu L., Zhao M., Wang T.-N., et al. Characterization and dye decolorization ability of an alkaline resistant and organic solvents tolerant laccase from Bacillus licheniformis LS04. Bioresource Technology. 2012;115:35–40. doi: 10.1016/j.biortech.2011.07.111. [ DOI ] [ PubMed ] [ Google Scholar ]
- 28. Madhavi V., Lele S. Laccase: properties and applications. Bio Resources. 2009;4(4):1694–1717. [ Google Scholar ]
- 29. Rajeswari M., Bhuvaneswari V. Production of extracellular laccase from the newly isolated Bacillus sp. PK4. African Journal of Biotechnology. 2016;15(34):1813–1826. [ Google Scholar ]
- 30. Akbar M. T., Habib A. M. An insight into the lignin peroxidase of Macrophomina phaseolina. Bioinformation. 2013;9(14):730–735. doi: 10.6026/97320630009730. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 31. Falade A. O., Nwodo U. U, Iweriebor B. C, Green E, Mabinya L. V, Okoh A. I. Lignin peroxidase functionalities and prospective applications. Microbiology Open. 2017;6 doi: 10.1002/mbo3.394.e00394 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 32. Wang X., Yao B., Su X. Linking enzymatic oxidative degradation of lignin to organics detoxification. International Journal of Molecular Sciences. 2018;19(11):p. 3373. doi: 10.3390/ijms19113373. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 33. Christian V. Mediator role of veratryl alcohol in the lignin peroxidase-catalyzed oxidative decolorization of Remazol Brilliant Blue R. Enzyme and Microbial Technology. 2005;36(2-3):327–332. doi: 10.1016/j.enzmictec.2004.09.006. [ DOI ] [ Google Scholar ]
- 34. Abdel-Hamid A., Solbiati J. O., Cann I. K. O. Insights into lignin degradation and its potential industrial applications. Advances in Applied Microbiology. 2013;82:p. 1. doi: 10.1016/B978-0-12-407679-2.00001-6. [ DOI ] [ PubMed ] [ Google Scholar ]
- 35. Abdel-Hamid A. M., Solbiati J. O., Cann I. K. O. Insights into lignin degradation and its potential industrial applications. Advances in Applied Microbiology. 2013;82:1–28. doi: 10.1016/b978-0-12-407679-2.00001-6. [ DOI ] [ PubMed ] [ Google Scholar ]
- 36. Cerniglia C. E. Biodegradation of polycyclic aromatic hydrocarbons. Current Opinion in Biotechnology. 1993;4(3):331–338. doi: 10.1016/0958-1669(93)90104-5. [ DOI ] [ Google Scholar ]
- 37. Shaheen R., Asgher M., Hussain F., Bhatti H. N. Immobilized lignin peroxidase from Ganoderma lucidum IBL-05 with improved dye decolorization and cytotoxicity reduction properties. International Journal of Biological Macromolecules. 2017;103:57–64. doi: 10.1016/j.ijbiomac.2017.04.040. [ DOI ] [ PubMed ] [ Google Scholar ]
- 38. Gold M. H., Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiological Reviews. 1993;57(3):605–622. doi: 10.1128/mmbr.57.3.605-622.1993. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 39. Bilal M., Asgher M. Dye decolorization and detoxification potential of Ca-alginate beads immobilized manganese peroxidase. BMC Biotechnology. 2015;15(1):p. 111. doi: 10.1186/s12896-015-0227-8. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 40. Bilal M., Asgher M., Iqbal M., Hu H., Zhang X. Chitosan beads immobilized manganese peroxidase catalytic potential for detoxification and decolorization of textile effluent. International Journal of Biological Macromolecules. 2016;89:181–189. doi: 10.1016/j.ijbiomac.2016.04.075. [ DOI ] [ PubMed ] [ Google Scholar ]
- 41. Chen M., Zeng G., Lai C., et al. Interactions of carbon nanotubes and/or graphene with manganese peroxidase during biodegradation of endocrine disruptors and triclosan. Chemosphere. 2017;184:127–136. doi: 10.1016/j.chemosphere.2017.05.162. [ DOI ] [ PubMed ] [ Google Scholar ]
- 42. Qin X., Zhang J., Zhang X., Yang Y. Induction, purification and characterization of a novel manganese peroxidase from Irpex lacteus CD2 and its application in the decolorization of different types of dye. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0113282.e113282 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 43. Mielgo I., Lopez C., Moreira M. T., Feijoo G., Lema J. M. Oxidative degradation of azo dyes by manganese peroxidase under optimized conditions. Biotechnology Progress. 2003;19(2):325–331. doi: 10.1021/bp020136w. [ DOI ] [ PubMed ] [ Google Scholar ]
- 44. Hirano T., Honda Y., Watanabe T., Kuwahara M. Degradation of bisphenol A by the lignin-degrading enzyme, manganese peroxidase, produced by the white-rot Basidiomycete, Pleurotus ostreatus. Bioscience, Biotechnology, and Biochemistry. 2000;64(9):1958–1962. doi: 10.1271/bbb.64.1958. [ DOI ] [ PubMed ] [ Google Scholar ]
- 45. Moon D.-S., Song H.-G. Degradation of alkylphenols by white rot fungus Irpex lacteus and its manganese peroxidase. Applied Biochemistry and Biotechnology. 2012;168(3):542–549. doi: 10.1007/s12010-012-9795-4. [ DOI ] [ PubMed ] [ Google Scholar ]
- 46. Xu H., Guo M. Y., Gao Y. H., Bai X. H., Zhou X. W. Expression and characteristics of manganese peroxidase from Ganoderma lucidum in Pichia pastoris and its application in the degradation of four dyes and phenol. BMC Biotechnology. 2017;17(1):19–12. doi: 10.1186/s12896-017-0338-5. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 47. Asgher M., Bhatti H. N., Ashraf M., Legge R. L. Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation. 2008;19(6):771–783. doi: 10.1007/s10532-008-9185-3. [ DOI ] [ PubMed ] [ Google Scholar ]
- 48. Knop D., Levinson D., Makovitzki A., et al. Limits of versatility of versatile peroxidase. Applied and Environmental Microbiology. 2016;82(14):4070–4080. doi: 10.1128/aem.00743-16. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 49. Gao X., Huang S., Dong P., et al. Horseradish peroxidase (HRP): a tool for catalyzing the formation of novel bicoumarins. Catalysis Science & Technology. 2016;6(10):3585–3593. doi: 10.1039/c5cy01682g. [ DOI ] [ Google Scholar ]
- 50. Bretz R. R., de Castro A. A., Lara Ferreira I. F., Ramalho T. C., Silva M. C. Experimental and theoretical affinity and catalysis studies between halogenated phenols and peroxidases: understanding the bioremediation potential. Ecotoxicology and Environmental Safety. 2020;202 doi: 10.1016/j.ecoenv.2020.110895.110895 [ DOI ] [ PubMed ] [ Google Scholar ]
- 51. Veitch N. C. Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry. 2004;65(3):249–259. doi: 10.1016/j.phytochem.2003.10.022. [ DOI ] [ PubMed ] [ Google Scholar ]
- 52. Köller G., Möder M., Czihal K. Peroxidative degradation of selected PCB: a mechanistic study. Chemosphere. 2000;41(12):1827–1834. doi: 10.1016/s0045-6535(00)00132-6. [ DOI ] [ PubMed ] [ Google Scholar ]
- 53. Tonegawa M., Dec J., Bollag J.-M. Use of additives to enhance the removal of phenols from water treated with horseradish and hydrogen peroxide. Journal of Environment Quality. 2003;32(4):1222–1227. doi: 10.2134/jeq2003.1222. [ DOI ] [ PubMed ] [ Google Scholar ]
- 54. Zheng W., Colosi L. M. Peroxidase-mediated removal of endocrine disrupting compound mixtures from water. Chemosphere. 2011;85(4):553–557. doi: 10.1016/j.chemosphere.2011.06.064. [ DOI ] [ PubMed ] [ Google Scholar ]
- 55. Mohan S. V., Prasad K. K., Rao N. C., Sarma P. N. Acid azo dye degradation by free and immobilized horseradish peroxidase (HRP) catalyzed process. Chemosphere. 2005;58:1097–1105. doi: 10.1016/j.chemosphere.2004.09.070. [ DOI ] [ PubMed ] [ Google Scholar ]
- 56. Onder S., Celebi M., Altikatoglu M., Hatipoglu A., Kuzu H. Decolorization of naphthol blue black using the horseradish peroxidase. Applied Biochemistry and Biotechnology. 2011;163(3):433–443. doi: 10.1007/s12010-010-9051-8. [ DOI ] [ PubMed ] [ Google Scholar ]
- 57. Li J., Chen X., Xu D., Pan K. Immobilization of horseradish peroxidase on electrospun magnetic nanofibers for phenol removal. Ecotoxicology and Environmental Safety. 2019;170:716–721. doi: 10.1016/j.ecoenv.2018.12.043. [ DOI ] [ PubMed ] [ Google Scholar ]
- 58. Alshawafi W. M., Aldhahri M., Almulaiky Y. Q., et al. Immobilization of horseradish peroxidase on PMMA nanofibers incorporated with nanodiamond. Artificial Cells, Nanomedicine, and Biotechnology. 2018;46(3):S973–S981. doi: 10.1080/21691401.2018.1522321. [ DOI ] [ PubMed ] [ Google Scholar ]
- 59. Šekuljica N. Ž. Immobilization of horseradish peroxidase onto kaolin. Bioprocess and Biosystems Engineering. 2016;39(3):461–472. doi: 10.1007/s00449-015-1529-x. [ DOI ] [ PubMed ] [ Google Scholar ]
- 60. Singh S., Malhotra S., Mukherjee P., et al. Peroxidases from an invasive Mesquite species for management and restoration of fertility of phenolic-contaminated soil. Journal of Environmental Management. 2020;256 doi: 10.1016/j.jenvman.2019.109908.109908 [ DOI ] [ PubMed ] [ Google Scholar ]
- 61. Krainer F. W., Glieder A. An updated view on horseradish peroxidases: recombinant production and biotechnological applications. Applied Microbiology and Biotechnology. 2015;99(4):1611–1625. doi: 10.1007/s00253-014-6346-7. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 62. Lamb S. B., Lamb D. C., Kelly S. L., Stuckey D. C. Cytochrome P450 immobilisation as a route to bioremediation/biocatalysis. FEBS Letters. 1998;431(3):343–346. doi: 10.1016/s0014-5793(98)00771-6. [ DOI ] [ PubMed ] [ Google Scholar ]
- 63. Sher S., Rehman A. Use of heavy metals resistant bacteria-a strategy for arsenic bioremediation. Applied Microbiology and Biotechnology. 2019;103(15):6007–6021. doi: 10.1007/s00253-019-09933-6. [ DOI ] [ PubMed ] [ Google Scholar ]
- 64. Lamb D. C., Kelly D. E., Masaphy S., Jones G. L., Kelly S. L. Engineering of heterologous cytochrome P450 in Acinetobacter sp.: application for pollutant degradation. Biochemical and Biophysical Research Communications. 2000;276(2):797–802. doi: 10.1006/bbrc.2000.3541. [ DOI ] [ PubMed ] [ Google Scholar ]
- 65. Kumar S. Engineering cytochrome P450 biocatalysts for biotechnology, medicine and bioremediation. Expert Opinion on Drug Metabolism & Toxicology. 2010;6(2):115–131. doi: 10.1517/17425250903431040. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 66. Pinto É. S. M., Dorn M., Feltes B. C. The tale of a versatile enzyme: alpha-amylase evolution, structure, and potential biotechnological applications for the bioremediation of n-alkanes. Chemosphere. 2020;250 doi: 10.1016/j.chemosphere.2020.126202.126202 [ DOI ] [ PubMed ] [ Google Scholar ]
- 67. Sakaki T., Yamamoto K., Ikushiro S. Possibility of application of cytochrome P450 to bioremediation of dioxins. Biotechnology and Applied Biochemistry. 2013;60(1):65–70. doi: 10.1002/bab.1067. [ DOI ] [ PubMed ] [ Google Scholar ]
- 68. Sakaki T. Practical application of cytochrome P450. Biological and Pharmaceutical Bulletin. 2012;35(6):844–849. doi: 10.1248/bpb.35.844. [ DOI ] [ PubMed ] [ Google Scholar ]
- 69. Rylott E. L., Jackson R. G., Sabbadin F., et al. The explosive-degrading cytochrome P450 XplA: biochemistry, structural features and prospects for bioremediation. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2011;1814(1):230–236. doi: 10.1016/j.bbapap.2010.07.004. [ DOI ] [ PubMed ] [ Google Scholar ]
- 70. Cobbett C., Goldsbrough P. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annual Review of Plant Biology. 2002;53(1):159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [ DOI ] [ PubMed ] [ Google Scholar ]
- 71. Kang S. H., Singh S., Kim J.-Y., Lee W., Mulchandani A., Chen W. Bacteria metabolically engineered for enhanced phytochelatin production and cadmium accumulation. Applied and Environmental Microbiology. 2007;73(19):6317–6320. doi: 10.1128/aem.01237-07. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 72. Pratush A., Kumar A., Hu Z. Adverse effect of heavy metals (As, Pb, Hg, and Cr) on health and their bioremediation strategies: a review. International Microbiology. 2018;21(3):97–106. doi: 10.1007/s10123-018-0012-3. [ DOI ] [ PubMed ] [ Google Scholar ]
- 73. Kapahi M., Sachdeva S. Bioremediation options for heavy metal pollution. Journal of Health and Pollution. 2019;9(24) doi: 10.5696/2156-9614-9.24.191203.191203 [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 74. Yamamura S., Amachi S. Microbiology of inorganic arsenic: from metabolism to bioremediation. Journal of Bioscience and Bioengineering. 2014;118(1):1–9. doi: 10.1016/j.jbiosc.2013.12.011. [ DOI ] [ PubMed ] [ Google Scholar ]
- 75. Ojuederie O. B., Babalola O. O. Microbial and plant-assisted bioremediation of heavy metal polluted environments: a review. International Journal of Environmental Research and Public Health. 2017;14(12) doi: 10.3390/ijerph14121504. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 76. Naik M. M., Dubey S. K. Lead resistant bacteria: lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicology and Environmental Safety. 2013;98:1–7. doi: 10.1016/j.ecoenv.2013.09.039. [ DOI ] [ PubMed ] [ Google Scholar ]
- 77. Rahman Z., Singh V. P. Bioremediation of toxic heavy metals (THMs) contaminated sites: concepts, applications and challenges. Environmental Science and Pollution Research. 2020;27(22):27563–27581. doi: 10.1007/s11356-020-08903-0. [ DOI ] [ PubMed ] [ Google Scholar ]
- 78. Teeling H., Cypionka H. Microbial degradation of tetraethyl lead in soil monitored by microcalorimetry. Applied Microbiology and Biotechnology. 1997;48(2):275–279. doi: 10.1007/s002530051050. [ DOI ] [ PubMed ] [ Google Scholar ]
- 79. Azimi S., Moghaddam M. S. Effect of mercury pollution on the urban environment and human health. Environment and Ecology Research. 2013;1(1):12–20. doi: 10.13189/eer.2013.010102. [ DOI ] [ Google Scholar ]
- 80. Mahbub K. R., Bahar M. M., Labbate M., et al. Bioremediation of mercury: not properly exploited in contaminated soils! Applied Microbiology and Biotechnology. 2017;101(3):963–976. doi: 10.1007/s00253-016-8079-2. [ DOI ] [ PubMed ] [ Google Scholar ]
- 81. Singh S., Kumar V. Mercury detoxification by absorption, mercuric ion reductase, and exopolysaccharides: a comprehensive study. Environmental Science and Pollution Research. 2020;27(22):27181–27201. doi: 10.1007/s11356-019-04974-w. [ DOI ] [ PubMed ] [ Google Scholar ]
- 82. Xu J., Bravo A. G., Lagerkvist A., Bertilsson S., Sjöblom R., Kumpiene J. Sources and remediation techniques for mercury contaminated soil. Environment International. 2015;74:42–53. doi: 10.1016/j.envint.2014.09.007. [ DOI ] [ PubMed ] [ Google Scholar ]
- 83. Velásquez-Riaño M., Benavides-Otaya H. D. Bioremediation techniques applied to aqueous media contaminated with mercury. Critical Reviews in Biotechnology. 2016;36(6):1124–1130. doi: 10.3109/07388551.2015.1100156. [ DOI ] [ PubMed ] [ Google Scholar ]
- 84. Tadishetty Hanumanth Rao S., Papathoti N. K., Gundeboina R., Mohamed Y. K., Mudhole G. R., Bee H. Hexavalent chromium reduction from pollutant samples by achromobacter xylosoxidans SHB 204 and its kinetics study. Indian Journal of Microbiology. 2017;57(3):292–298. doi: 10.1007/s12088-017-0654-4. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 85. Chardin B., Giudici-Orticoni M.-T., De Luca G., Guigliarelli B., Bruschi M. Hydrogenases in sulfate-reducing bacteria function as chromium reductase. Applied Microbiology and Biotechnology. 2003;63(3):315–321. doi: 10.1007/s00253-003-1390-8. [ DOI ] [ PubMed ] [ Google Scholar ]
- 86. Gonzalez C. F., Ackerley D. F., Lynch S. V., Matin A. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. Journal of Biological Chemistry. 2005;280(24):22590–22595. doi: 10.1074/jbc.m501654200. [ DOI ] [ PubMed ] [ Google Scholar ]
- 87. Ackerley D. F., Gonzalez C. F., Park C. H., Blake R., Keyhan M., Matin A. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Applied and Environmental Microbiology. 2004;70(2):873–882. doi: 10.1128/aem.70.2.873-882.2004. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
- View on publisher site
- PDF (2.5 MB)
- Collections
Similar articles
Cited by other articles, links to ncbi databases.
- Download .nbib .nbib
- Format: AMA APA MLA NLM
Add to Collections
REVIEW article
Bioremediation of environmental wastes: the role of microorganisms.

- 1 Food Security and Safety Focus Area, Faculty of Natural and Agricultural Sciences, North-West University, Mmabatho, South Africa
- 2 Department of Biological Sciences, Kings University, Ode-Omu, Nigeria
- 3 Environmental Pollution Science and Technology, (ENPOST), Ido-Ijesha, Ilesha, Nigeria
The growing rate of urbanization and industrialization has led to an increase in several types of pollution caused by the release of toxic chemicals to the environment. This is usually perpetuated by the manufacturing industry (e.g. detergent and dye), agricultural sectors (e.g. fertilizers and pesticides), mining industry (e.g. cyanide and sulphuric acid) and construction companies (e.g. cement and metals). These pollutants have adverse effects on the health of plants, animals, and humans. They also lead to the destruction of the microbial population in both aquatic and the terrestrial regions, and hence, have necessitated the need for remediation. Although different remediation methods, such as the physical and chemical methods, have been adopted for years, however, the drawbacks and challenges associated with them have promoted the use of an alternative which is bioremediation. Bioremediation involves using biological agents such as plants and microbes to remove or lessen the effects of environmental pollutants. Of the two, microbes are more utilized primarily because of their rapid growth and ability to be easily manipulated, thus enhancing their function as agents of bioremediation. Different groups of bacteria, fungi and algae have been employed to clean up various environmental pollutants. This review discusses the types, mechanisms, and factors affecting microbial bioremediation. It also recommends possible steps that could be taken to promote the use of microbes as bioremediation agents.
1 Introduction
The rise of urbanization and industrialization, has left the environment exposed to numerous pollutants which are toxic to living things. Pollutants arising from different industrial processes are major sources of pollution to the soil and aquatic environment. Different types and quantities of heavy metals are released during the industrial production process and as effluents after further industrial production. For instance, the wastewater from dye-producing companies are associated with antimony, chromium and mercury ( Methneni et al., 2021 ). The application of fertilizers, pesticides and herbicides in the agricultural sector generates pollutants that include aluminium, copper, zinc, nickel, lead and arsenic to the environment ( Ayilara et al., 2020 ; Prabagar et al., 2021 ). Similarly, untreated pollutants from wastewaters of the agri-food industries disposed into river canals and other waterbodies have harmful effects on the environment ( Siric et al., 2022a ; AL-Huqail et al., 2022 ). Crude oil also serves as a major environmental pollutant particularly through pipeline vandalization, transportation leakage, and/or accidental spillage ( Ogunlaja et al., 2019 ). During mining, some chemicals such as lead, arsenic, cadmium, and copper which are toxic to the immediate environment are released ( Liu et al., 2020 ). Some other environmentally toxic chemicals including but not limited to cyanide and sulphuric acid are used during the mining process. ( Ayangbenro et al., 2018 ; Orlovic-Leko et al., 2022 ). Equally, other industrial wastes such as those produced in cement-making industries release zinc, copper and cadmium and can be found in the top soils ( Jafari et al., 2019 ). Chromium and lead from pharmaceutical effluents ( Kumari and Tripathi, 2020 ), plastics containing lead, manganese, iron, copper, chromium, silver, cadmium, antimony and mercury all pollute water ( Zhou et al., 2019 ). In addition, copper, arsenic, mercury, chromium, lead, nickel, cadmium and zinc from the coal industry serve as environmental pollutant ( Sun et al., 2019 ). These heavy metals are very toxic to aquatic and terrestrial habitats and their inhabitants. In humans, mercury, cadmium and lead alters the central nervous system, especially in infants, while lead results in liver and kidney dysfunction, cardiovascular diseases, malfunctioning of the reproductive and immune system ( Zwolak et al., 2019 ; Fashola et al., 2020a ; Fashola et al., 2020b ; Ayangbenro and Babalola, 2020 ). Cadmium causes cancers, skeletal disorders, neurotoxic and nefrotoxic complexities, and dysfunction of the reproductive system ( Zwolak et al., 2019 ; Fashola et al., 2020a ; Fashola et al., 2020b ; Ayangbenro and Babalola, 2020 ). Wastes containing heavy metals are often improperly disposed into soil and water environments. When disposed into water bodies, they can lead to the death of fishes, and other aquatic inhabitants, otherwise, they are biomagnified and cause chronic diseases in humans and animals. Therefore, there is need for the remediation of these pollutants using physical, chemical, or biological methods. The physical and chemical methods have been used for years but they come with their drawbacks which include the need for an expert and special equipment for the chemical bioremediation procedure while the physical bioremediation procedure is expensive ( Mahmood et al., 2021 ). This has called for the need for a better alternative which is the biological remediation (Bioremediation). Bioremediation is a most efficient, eco-friendly and cost effective technology for the transformation of contaminants ( Sonune, 2021 ). Biological remediation can be carried out using both plants and microorganism, nonetheless, plants take a longer time to grow and cannot be easily manipulated like the microbes which makes the microbes more preferable ( Hussain et al., 2022 ). In addition, microbes mitigates heavy metals and improve soil fertility and plant development ( Chaudhary et al., 2023b ). Hence, this review discusses the types, mechanism, challenges as well as the factors affecting microbial bioremediation, with recommendation on how to enhance the use of microbes in aquatic and terrestrial bioremediation.
2 Different pollutants and their toxicity on living things
Exposure of humans to air pollutants can cause developmental disorders, respiratory problems, cancers, cardiovascular diseases, and other health issues ( Table 1 ). For instance, it has been reported that exposure to particulate matter in the air was associated with an increased risk of premature death in humans ( Pope et al., 2019 ). Nitrogen oxides produced by combustion processes, are significant air pollutants. They irritate the respiratory system, cause cough, shortness of breath, and exacerbate asthma ( Zhao et al., 2020 ). Equally, Sulfur dioxide, produced by burning fossil fuels, can cause respiratory and cardiovascular diseases, including bronchoconstriction, shortness of breath, and coughing. A recent study found that exposure to sulfur dioxide was associated with increased mortality from respiratory diseases in China ( Luo et al., 2015 ). Volatile organic compounds (VOCs), emitted by various sources, including paints, cleaning products, and vehicle emissions, can cause eye, nose, and throat irritation, headaches, nausea, and dizziness. Some VOCs (such as benzene) are also carcinogenic, and are associated with an increased risk of leukemia ( Bala et al., 2021 ). Water pollutants which include pesticides, heavy metals, and organic compounds are sometimes ingested by humans either directly or indirectly (through the consumption of aquatic animals). These pollutants can cause various health problems, including cancer, neurological disorders, and reproductive issues. It has been reported that exposure to heavy metals result in a higher risk of hypertension and kidney damage in humans ( Wu et al., 2018 ; Rai et al., 2019 ).
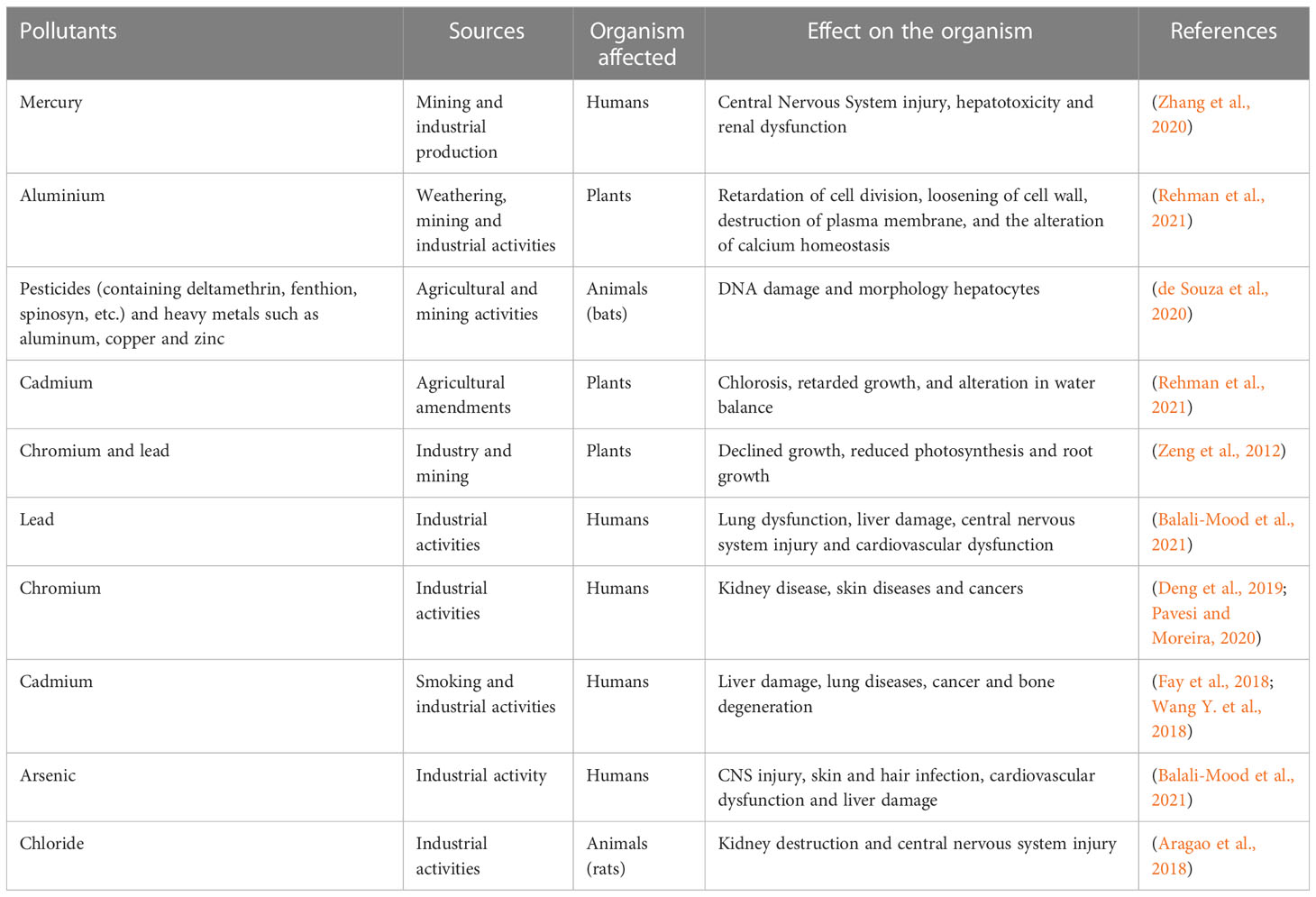
Table 1 Effect of pollutants on living things.
Similarly, different animal diseases are caused by pollutants. Exposure to particulate matter (PM) can cause inflammation and damage to the respiratory system of animals, leading to respiratory diseases such as chronic obstructive pulmonary disease (COPD) and asthma ( Manisalidis et al., 2020 ). When animals consume water contaminated with heavy metals, pesticides, and pharmaceuticals, it leads to reproductive disorders, liver damage, and cancer ( Hitt et al., 2023 ). Nitrogen dioxide when present in the environment, reduces the growth of plants and the yield of crops while sulfur dioxide causes acid rain and acidification ( Manisalidis et al., 2020 ). An impairment in the photosynthetic rhythm and metabolism is observed in plants exposed to ozone ( Zuhara and Isaifan, 2018 ). In the aquatic environment, eutrophication occurs when there is a high concentration of nitrogen availability. This leads to algal bloom and cause death and disequilibration in the diversity of fish ( Zuhara and Isaifan, 2018 ).
2.1 Types of remediation
There are different types of remediation, namely the physical, chemical and biological techniques. The physical remediation involves the use of skimmers, sorbent materials and booms. Boom is a physical barrier made of materials that absorbs oil pollutants and prevents it from spreading before a further remediation procedure is carried out ( Vocciante et al., 2019 ) ( Figure 1 ). Skimmers and sorbents are methods that are further used to absorb and adsorb pollutants after booms ( Kumari et al., 2019 ). The major challenge associated with the use of bloom remediation technique is that it is dependent on the buoyancy and roll response. When the boom is buoyant, it floats and remains longer on the water surface. The roll response refers to the torque required to rotate the bloom from its vertical position. That is, an increased roll response results in a higher remediation process ( Dhaka and Chattopadhyay, 2021 ).
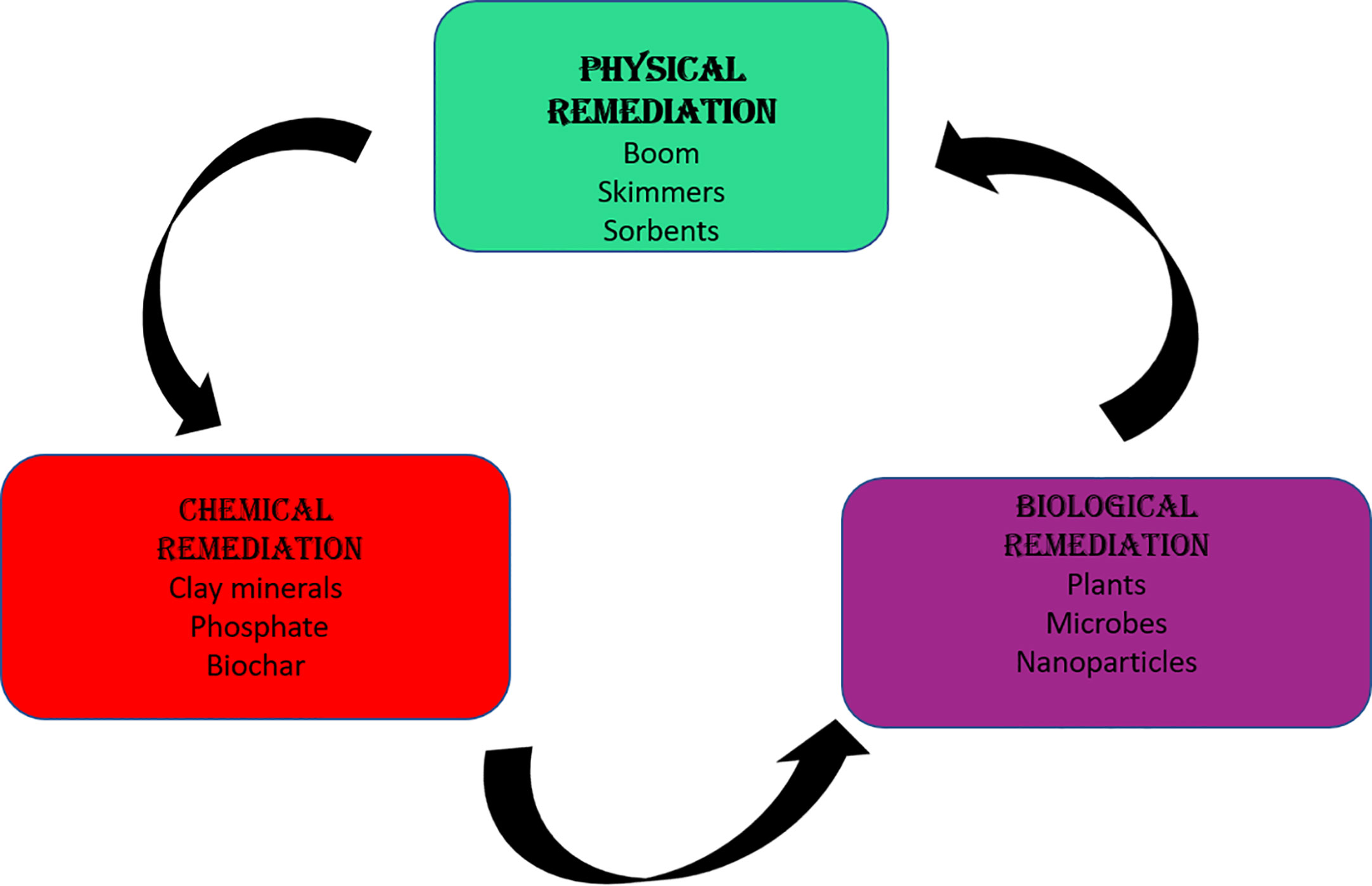
Figure 1 Types of bioremediations.
Chemical remediation is the process of adding chemicals such as clay minerals, phosphate, biochar, aluminum salts, silicocalcium materials, and sulfide to stabilize and remove heavy metals from the environment. The mechanism behind the use of these chemicals include adsorption, reduction, oxidation, complexation, precipitation and ion exchange ( Xu et al., 2022 ). Chemical remediation technique is an easy, simple, and rapid technique; however, the chemical used can also serve as a source of environmental pollution ( Xu et al., 2022 ) ( Figure 1 ).
Bioremediation is another method of pollution treatment, it is a sustainable, affordable and safe remediation technique ( Kumar A. et al., 2021 ; Kumar G. et al., 2021 ; Patel A. K. et al., 2022 ). The technology involves the use of organics such as plants and microbes. The viability of this method depends on the nature, location and level of pollution ( Patel A. K. et al., 2022 ). Microbes on the other hand have proved to be efficient in the remediation of environmental pollutants. They are preferred to plants in remediation, this is due to their ease of growth, rapid growth period and easy manipulation. It is therefore necessary to improve the use of microbes as agent of bioremediation to promote a sustainable environment.
3 Different microbes used as bioremediation agents
Microorganisms can convert toxic elements into water, carbon dioxide, and other less toxic compounds, which are further degraded by other microbes in a process referred to as mineralization ( Mahmoud, 2021 ; Kumar G. et al., 2022 ). Bioremediation can be carried out using bacteria, fungi, algae, etc. ( Table 2 ). Microbes are ubiquitous in nature, and they utilize a wide range of substrates as carbon source; hence, they are found in unusual environments where they can absorb a wide range of pollutants ( Kour et al., 2022 ). Also, their ability to survive in odd environments promote their efficiency. For example the acidophiles survive in acidic environments, the psychrophiles thrive in cold climates and the halophiles survive in saline region ( Perera and Hemamali, 2022 ).
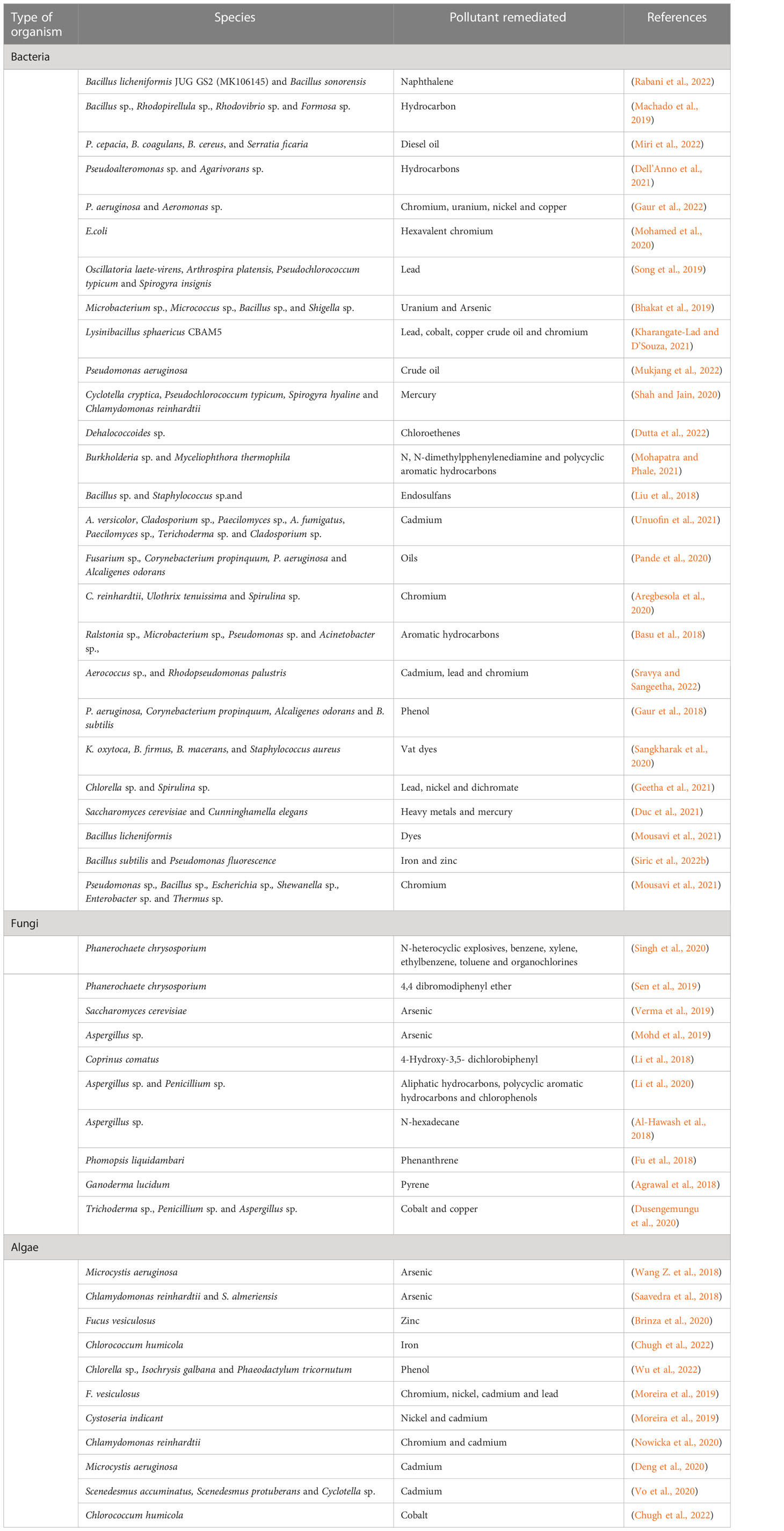
Table 2 Different microbes used in bioremediation.
4 Mechanisms of microbial bioremediation
Microbes can remove pollutants from the environment using different mechanisms. These mechanisms can be placed into two broad categories namely immobilization and mobilization ( Ndeddy Aka and Babalola, 2016 ; Verma and Kuila, 2019 ). Mobilization process involves, enzymatic oxidation, bioleaching, biostimulation, bioaugmentation and enzymatic reduction procedure. On the other hand, immobilization includes bioaccumulation, complexation, biosorption, and precipitation (solidification) ( Tak et al., 2012 ; Ayangbenro et al., 2019 ). During mineralization, microbes help transform pollutants into end products such as carbon dioxide and water or other intermediate metabolic substances. Similarly, immobilization is the conversion of compounds into a form where it will be unavailable in the environment. For instance, the conversion of nitrate nitrogen into organic nitrogen ( Pratush et al., 2018 ). The method is usually utilized for the bioremediation of heavy metals, especially in highly contaminated environments.
Immobilization can be carried out using the in-situ and the ex-situ methods ( Pratush et al., 2018 ). The ex-situ process involves the removal of polluted soils from the site of pollution to another location where it would undergo a microbial process to immobilize the metal ions responsible for the contamination ( Ayangbenro and Babalola, 2017 ). On the other hand, in the in-situ procedure, the pollution is treated on site ( Cao et al., 2020 ). Microbes such as E. asburiae and B. cereus have been reported to be involved in immobilization of heavy metals which pollute the environment ( Fashola et al., 2020a ). During microbial bioremediation, microbes protect themselves from toxic compounds by forming hydrophobic or solvent efflux pump that protects the outer membrane of the cell ( Verma and Kuila, 2019 ).
4.1 Enzymatic oxidation
Enzymatic oxidation is the process of oxidizing pollutant compounds from a higher oxidation state to a lower one, during which heavy metals lose an electron and become less toxic. This process utilizes an enzyme (oxidoreductase) released by the microbes involved. This method is highly effective in the remediation of dyes, phenols, and other pollutants which are not easily degraded by bacteria ( Unuofin et al., 2019 ). The oxidative enzymes form radicals which can be broken down into different fractions, eventually forming compounds with high molecular weight ( Unuofin et al., 2019 ). An example of an oxidoreductase enzyme is laccase, which catalyzes the oxidation of aromatic amines ( Gangola et al., 2018 ). Other examples are phenols and polyphenols, which cause the reduction of molecular oxygen to water ( Kushwaha et al., 2018 ; Sahay, 2021 ). Laccase production has been reported in Pycnoporus sp. and Leptosphaerulina sp. where it was outlined to degrade heavy metals ( Copete-Pertuz et al., 2018 ; Tian et al., 2020 ).
4.2 Enzymatic reduction
This process is the opposite of enzymatic oxidation, here, the pollutants are converted to a reduced oxidized state where they become insoluble. Obligate and facultative anaerobes carry out the process; this method is effective in the bioremediation of compounds such as polychlorinated dibenzo-p-dioxins and dibenzofurans ( Zacharia, 2019 ). Equally, chrome reductase catalyzes the reduction of hexavalent chromium to trivalent chromium, and azoreductase reduces the azo compounds by cleaving to azo bonds ( Saxena et al., 2020 ). Much more research is needed to unravel other organisms which are capable of bioremediating pollutants in the environment.
4.3 Bioaugmentation
Microorganisms are specially added to polluted sites to feed on toxic pollutants in a process referred to as bioaugmentation. It is a very effective, rapid and cost-effective method of bioremediation ( Mahmoud, 2021 ). External microbes are added to polluted sites to augment the resident microbes. In other cases, it could also involve the isolation and genetic modification of microbes from the site of pollution before returning them to the same site for remediation. Genetic manipulation of resident microbes of polluted sites is carried out because the organisms may naturally not be capable of degrading the pollutant present at a site, and hence are modified to enhance their ability. In some other cases, non-resident microbes are added to polluted areas to promote the degradation of pollutants. The effectiveness of these new strains depends on some factors, which include the ability to compete with the resident microbes and the ability to adapt to the new environment ( Fashola et al., 2016 ; Ayangbenro and Babalola, 2017 ; Goswami et al., 2018 ; Babalola et al., 2019 ). Burkholderia sp. FDS-1 which was added to a polluted site, has been reported to degrade nitrophenolic compound present in pesticides polluted soil to a less toxic form at a slightly acidic pH and a temperature of about 30° C ( Goswami et al., 2018 ; Ojuederie et al., 2021 ) ( Table 3 ).
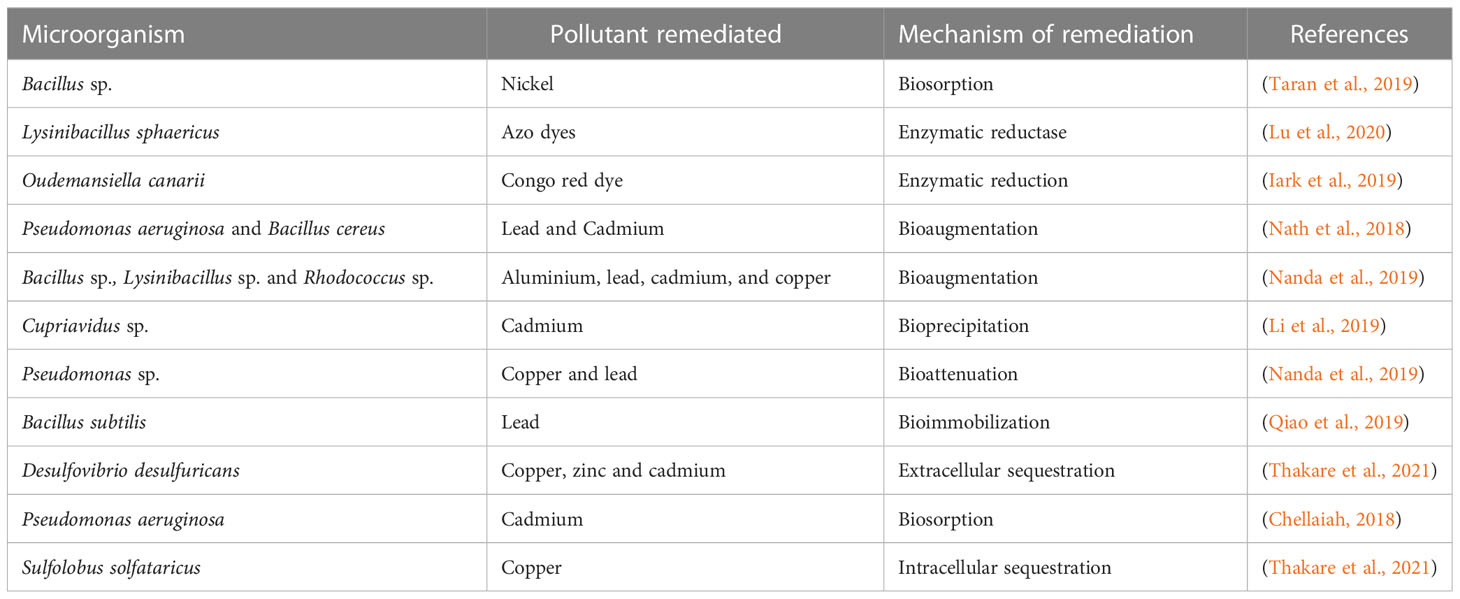
Table 3 Mechanism of Bioremediation.
4.4 Biostimulation
Biostimulation is the addition of nutrients (such as nitrogen, potassium, phosphorus), metabolites, electron donors, enzymes, electron acceptors, biosurfactants, etc., which are limiting to the soil to enhance the activity of the resident microbes and increase the remediation process ( Ojuederie and Babalola, 2017 ; Ayangbenro and Babalola, 2018 ). It is an affordable, environmentally friendly and efficient process ( Goswami et al., 2018 ). Compared to the bioaugmentation method, the biostimulation method is preferable because indigenous microbes are more competitive than the introduced ones ( Sayed et al., 2021 ), and this method helps to maintain the natural microbial diversity balance of the environment. Nivetha et al. (2022) reported the effectiveness of Bacillus sp., Rhodococcus sp., Staphylococcus sp., Klebsiella sp., Pseudomonas sp., and Citrobacter sp. in bioremediation of heavy metals through the biostimulation technique. Unfortunately, as effective as this method of bioremediation may be, it could lead to some other environmental complications, including eutrophication due to the excess nutrient present in the environment. Also, if the sources of the nutrients are chemicals (synthetic), they can serve as a source of pollution to the environment defeating the initial purpose of bioremediation ( Table 3 ).
4.5 Bioleaching
Bioleaching is the process of utilizing acidophilic microbes to promote the solubilization of heavy metals which are in a solid state from the sediment matrix. The process is particularly useful for iron or sulfur pollutants ( Sun et al., 2021 ; Bhandari et al., 2023 ). Therefore, iron- or sulfur-oxidizing bacteria are majorly recruited for this process; examples of such organisms are A. thiooxidans , Aspergillus sp., Mucor sp., Penicillium sp., Cladosporium sp. and Rhizopus sp. ( Medfu Tarekegn et al., 2020 ). These microbes create an acid environment and solubilize heavy metals in an immobilized state, into an aqueous solution ( Medfu Tarekegn et al., 2020 ).
4.6 Biosorption
This is the adsorption of heavy metals from pollutants through proton and ion displacement, complexation, chelation and physical interaction with electrostatic forces ( Mahmoud, 2021 ). It involves the removal of contaminants from solutions as a result of the outer cell shield of bacteria, fungi and algae which are bioremediation agents. Generally, metals are linked through the active groups of the compounds which exist at the cells surface layer. This results in the transfer of ion between metal cations and the negatively charged active group potentials present at the outer part of the microorganism structure. Rhodococcus erythropolis , Streptomyces sp. K11, and Bacillus anthracis have been reported to be capable of bioremediation through the biosorption process ( Mathew and Krishnamurthy, 2018 ; Baltazar et al., 2019 ; Sedlakova-Kadukova et al., 2019 ). Oftentimes, heavy metal pollutants (e.g., gold, zinc and copper) have some economic importance and are very useful in industrial processes. Hence, the ability of the compounds to be recovered through a process called desorption (using the solution of weak mineral solution or chelating compounds), which is a reversible step in biosorption makes it a good process ( Medfu Tarekegn et al., 2020 ).
Complexation involves using ligand to form a complex with inorganic metals, which are pollutants in the environment, especially solid wastes ( Ayangbenro and Babalola, 2017 ). Complexation is carried out mainly through different agents, namely the high molecular weight ligands, siderophores and toxic metal-binding compounds as well as the low-molecular weight organic acids (alcohols, tricarboxylic acids and citric acids) ( Pratush et al., 2018 ). Complexation occurs when extracellular polymeric substances, found on the surfaces of microbes interact with heavy metals which pollute the environment ( Xie et al., 2020 ). Xiao et al. (2019) reported the removal of copper (II) oxide and hexavalent chromium from wastewater using biochar in a mechanism which includes complexation. The organisms that have been reported to be involved in complexation include Rhodobacter blasticus ( Bai et al., 2019 ) and B.lichenformisis ( Wang et al., 2019 ).
When microbes are exposed to a polluted environment where there is iron-deficiency, they produce siderophores which are iron chelators. The siderophores have binding groups such as hydroxamate, catecholate and phenolates that form complexes with heavy metals and increase their solubility ( Khan et al., 2018 ). Siderophores are capable of producing reactive oxygen species, which also enhance their function as bioremediation agents for organic contaminants ( Albelda-Berenguer et al., 2019 ). Cyanobacteria have been reported to be effective as bioremediation agents due to the production of siderophores; for example, they are capable of bioremediating complex compounds like polythene and are capable of producing different types of siderophores, which include the anachelin, synechobactin and schizokinen ( Arstol and Hohmann-Marriott, 2019 ; Sarmah and Rout, 2020 ) ( Table 3 ).
4.7 Bioaccumulation
Bioaccumulation refers to the process where the rate of absorption of a compound is more than the rate at which the compound is lost. This process leads to the (toxic) build-up of compounds in the intracellular portion of the microbes. ( Sharma et al., 2022a ). Heavy metals move across the membrane of microbes using different mechanisms such as carrier-mediated transport, protein channel and ion pumps ( Mir-Tutusaus et al., 2018 ). Many organisms have been reported to be very active in bioaccumulation of heavy metals. For example, Rhizopus arrhizus , bioremediates mercury, Pseudomonas putida , bioremediates cadmium and Aspergillus niger bioremediates thorium ( Sharma et al., 2022a ).
4.8 Precipitation
This is the conversion of heavy metals or pollutants into precipitates or crystals, resulting in a reduced toxicity level; this process can occur during the biogeochemical cycling to form deposing of metals (iron and manganese), mineralized manganese and silver as well as microfossils, due to the activity of enzymes and galactosis of secondary metabolites ( Sharma et al., 2022a ). For instance, sulfate-reducing bacteria are capable of converting organo-phosphate to ortho-phosphate when the pH is alkaline (i.e. above 7) ( Pratush et al., 2018 ). Similarly, Bacillus subtilis and Oceanobacillus indicireducens have also been reported to be associated with the precipitation of heavy metals in the environment ( Maity et al., 2019 ).
5 Factors affecting microbial bioremediation
The ability of microbes to bioremediate heavy metals is determined by different factors, which include the total metal ion concentration, redox potential, chemical forms of the metals, competition among microbes, pH, temperature, soil structure, presence of oxygen, moisture content, nature of the soil and the solubility of the heavy metal in water ( Medfu Tarekegn et al., 2020 ). At acidic pH, free ionic species are formed by heavy metals, leading to the availability of more protons which would saturate the binding site of the metals. The pH of an environment affects the structure of the pollutant and also determines the ability of the microbe to survive in such an environment; the optimum pH that enhances bioremediation falls between 6.5 and 8.5 ( Kharangate-Lad and D’Souza, 2021 ).
Microbes compete for carbon which is a limited resource and serve as an energy source for microbes. Therefore, the inherent ability of the microbes, which compete better to degrade heavy metal pollutant, would affect the biodegradation rate. In addition to carbon, microbes responsible for biodegradation also require nitrogen (N) and phosphorus (P), thus it is important to balance the C:N:P ratio to enhance the rate of biodegradation, in environment when these essential nutrients are limited. They can be added to increase microbial activities ( Bala et al., 2022 ). The type and population of microbes determine the rate and success of a bioremediation process, for instance in the laboratory, a strain of organism might successfully bioremediate a particular heavy metal, which becomes problematic in a field situation where a consortium of microbes would be needed ( Patel A. B. et al., 2022 ). The molecular nature, gene and enzyme induction, metabolite production, growth efficiency and survival rate affect the ability of individual microbes as bioremediation agents ( Kebede et al., 2021 ). In addition, the ionization of the cell wall’s chemical moieties, the configuration of the microbial cell wall and sorption site also affect the rate of microbial biodegradation ( Mahmoud, 2021 ).
The amount of moisture present in an environment affects the solubility of the heavy metals in water, as well as their availability, pH and osmotic pressure ( Medfu Tarekegn et al., 2020 ). At a high moisture content, the microbial biodegradation rate is very low. This might be a result of an anaerobic condition that is created, which prevents the survival of aerobic microbes. Also, at a low moisture content, microbes might not be able to survive; hence an optimum moisture content is required for a successful microbial biodegradation process. In the cold regions where only psychrophiles can survive, the rate of microbial degradation of heavy metals is slow. This is because metabolic activities are reduced as the microbial transport channels is freezed by the sub-zero water; the degradation of each compound also occurs at different temperature even though most bioremediation processes are favored by high temperature ( Ren et al., 2018 ; Bala et al., 2022 ; Sharma et al., 2022c ). At an increased temperature, the rate of heavy metal solubility is increased, which consequently increases their rate of availability as well as the rate of microbial biodegradation ( Mahmoud, 2021 ).
Similarly, the chemical structure, bioavailability, concentration, toxicity and stability of the metal or pollutant determines the rate at which microbial biodegradation takes place ( Kebede et al., 2021 ). For instance, heavy metals with a simple chemical structure and low concentration would be easier to be remediated by microbes compared to those with a complex chemical structure and high temperature. Cycloalkane compounds that are highly condensed as well as high molecular weight polymatic hydrocarbons (those containing four rings and above) are more difficult to degrade compared to the lighter polyhydrocarbons (anthracene, naphthalene and phenanthrene) and unbranched alkanes (alkanes with intermediate length of about C 10 –C 25 ) ( Kebede et al., 2021 ). Hence, in order of ascending degradation, the n-alkanes are more easily degraded compared to the branched alkanes, low molecular weight aromatics, high molecular weight hydrocarbons and the asphaltenes ( Imam et al., 2019 ). Biodegradation is carried out aerobically and anaerobically. The ability of an organism which degrades a particular nutrient to survive in such an environment depends on the nature of the organism ( Jacob et al., 2018 ). For example, oxygenase associated with organisms that are active in aerobic regions is only produced in the presence of oxygen.
Different soil parameters, including the soil region, moisture-holding capacity texture and particle size, affect the rate of microbial biodegradation ( Alvarez et al., 2020 ). There is a higher population and diversity of microbes at the top layer of the soil (0-10cm). This is due to the increased availability of oxygen and organic matter, which is the opposite of what happens in sediment soils ( Ndeddy Aka and Babalola, 2017 ). In soils with fine particles, such as clayey soils, hydrocarbon retention takes place more at the surface, which renders the nutrient of the soil and oxygen unavailable. Therefore the best soil texture that promotes increased microbial biodegradation is the well-drained soil which supports oxygen availability and inhabits more soil microbes ( Huang et al., 2019 ). The presence of salinity has an effect on the hydrocarbonoclastic activity of the halotolerant and halophilic microbes, and it also exposes the soil microbes to stress conditions. The osmotic pressure of microorganisms increases as the saline concentration of an environment increases. This has a direct negative impact on the metabolic activities, of the microbes as well as the transportation system and solubility of the heavy metals ( Imron et al., 2020 ; Kebede et al., 2021 ).
6 Microbial enzymes used in bioremediation
Different microbial enzymes have been reported to be helpful in the removal of pollutants (especially heavy metals) in the environment ( Verma and Kuila, 2019 ; Bhatt et al., 2021a ; Chaudhary et al., 2023a ) ( Table 4 ). Mechanisms such as elimination, oxidation, ring-opening and reduction are used by enzymes in bioremediation ( Bhandari et al., 2021 ). Different factors which include temperature, contact time, concentration and pH affect the potency of microbial enzymes ( Bhandari et al., 2021 ). Enzyme bioremediation is expensive and time-consuming and therefore cannot be used when there is an urgent need for bioremediation ( Narayanan et al., 2023 ). Equally, the stability and activity of the pollutants, affects the potency of the bioremediation process. It is difficult to determine and discover multiple sources of a particular type of enzyme which might make the procedure unsustainable ( Narayanan et al., 2023 ).
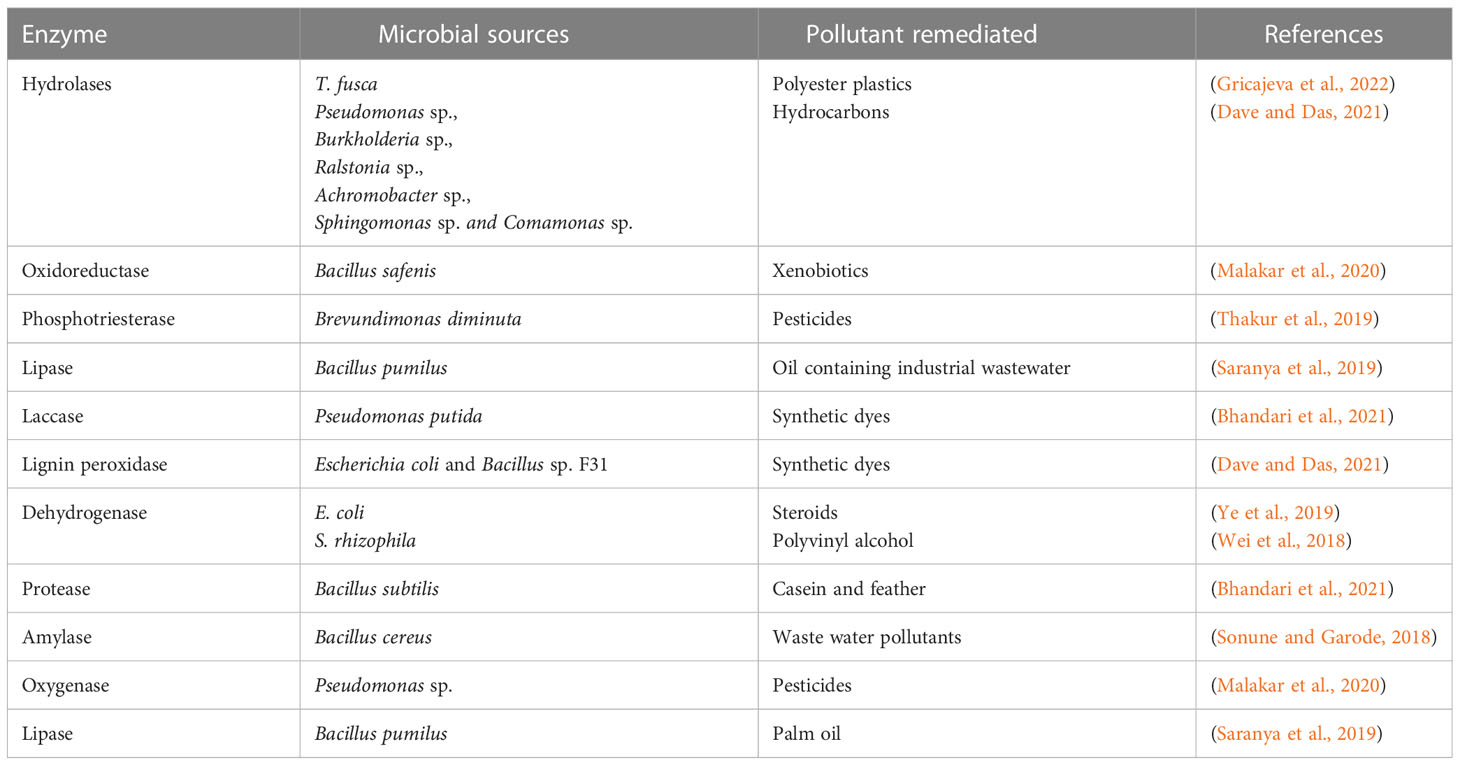
Table 4 Enzymes used in Microbial Bioremediation.
7 Molecular approaches for validating microbial remediation
Molecular mechanisms help to unravel the microbial metabolism, genes, nature, diversity and dynamics of microbes involved in microbial remediation. Diverse molecular mechanisms are utilized in the study of microbes used in bioremediation. Metabolic and protein profiling, sequencing as well as the use of advanced bioinformatics resources are particularly used to unravel the different groups of microbes and the factors affecting them in bioremediation process ( Sharma et al., 2022b ). On the other hand, conventional and culture-dependent molecular methods are also used in the monitoring of microbial communities during bioremediation. These methods include the use of terminal-restriction fragment (T-RF) length polymorphism, amplified ribosomal DNA restriction analysis, temperature gradient gel electrophoresis, randomly amplified polymorphic DNA analysis, length heterogeneity polymerase chain reaction, amplified fragment length polymorphisms, denaturing gradient gel electrophoresis, length heterogeneity polymerase chain reaction, automated ribosomal intergenic spacer analysis and single strand conformation polymorphism ( Bharagava et al., 2019 ).
Moreover, omics approaches such as transcriptomics, proteomics and metagenomics have greatly contributed in this field. Metagenomics involve the extraction of genomic DNA from all forms of life residing in a sample. Thereafter, the DNA will be fragmented, cloned, transformed and screened in the metagenome library ( Bharagava et al., 2019 ). The approaches to metagenomics include metabolomics, metatranscriptomics, fluxomics and metabolomics. Metatranscriptomics involve the use of metagenomic mRNA which unravel the function and expression of microbes present in a sample ( Mukherjee and Reddy, 2020 ). Metaproteomics involved the assessment of all the protein samples that comes from environmental samples ( Bargiela et al., 2015 ). Metabolomics is the identification and quantification of all the metabolites released into an environment ( Liu et al., 2022 ). Fluxomics refers to the different approaches used to study the rate of metabolic activities in a biological sample ( Kumar V. et al., 2022 ). More recently, the use of Next-Generation sequencing which is viewed as the most powerful technology for gene sequencing has become more popular ( Eisenhofer et al., 2019 ).
8 Other bioremediation metabolites produced by microbes
Microbes produce metabolites such as organic acids, biosurfactants and polymeric substances which are also used in bioremediation. Organic acids improve the bioavailability, mobility and solubility of metals; examples of organic acids include citric acids, malate and acetic acids ( Saha et al., 2021 ). Polymeric substances are beneficial in bioremediation by enhancing the phytostabilization of metals (through mobility), examples of polymeric substances include polyesters, polysaccharides and polyphosphates. Equally, biosurfactants which include viscosin, polymixin, glycoprotein and gramicidin help to solubilise, mobilise and increase the bioavailability of hydrophobic substrates ( Ojuederie and Babalola, 2017 ; Saha et al., 2021 ).
9 Recent advancements in microbial bioremediate
Lately, many improvements have been observed with the use of microbes as agents of bioremediation. Microbial glycoconjugates help to reduce the surface tension, increase the bioavailability, and create a solvent interface of organic pollutants. This helps to enhance the removal of the pollutants in the environment ( Bhatt et al., 2021b ). Atakpa et al. (2022) reported the use of microbial glycoconjugates from Scedosporium sp. and Acinetobacter sp. in the biodegradation of petroleum hydocarbons.
Microbial biofilms which consist of polysaccharides, extracellular DNAs and proteins are also lately used in the bioremediation of organic pollutants ( Sonawane et al., 2022 ). They are particularly used in the remediation of recalcitrant pollutants. The technology is presently being made better by improving on the quorum sensing, environmental factors and surface of adhesion ( Sonawane et al., 2022 ). In a research carried out by Andreasen et al. (2018) , it was revealed that Exiguobacterium profundum was able to significantly reduce the concentration of arsenic in synthetic wastewater after 48 hours of incubation.
Bioelectrochemical system is another emerging technology which combines the use of biological and electrochemical methods in the control of pollutants ( Ambaye et al., 2023 ). This technology helps to majorly remediate petroleum hydrocarbon pollutants and its efficiency depends mainly on the syntrophic and cooperative interactions between the members of the microbial groups involved ( Ambaye et al., 2023 ). Sharma et al. (2020) stated that Pseudomonas sp. , Ralstonia sp. , Rhodococcus sp., and Thauera sp. are capable of remediating phenanthrene from petroleum hydrocarbon polluted soils.
Nanotechnology is a thriving method of pollution control globally. Nanomaterials can be sourced from different sources which include the physical and chemical sources ( Shanmuganathan et al., 2019 ). The efficiency of nanoparticles as bioremediation agents is dependent on different factors such as the size, chemical nature, surface coating and shape of the nanoparticles ( Tan et al., 2018 ). Other factors such as the nature of the pollutants, type of media, temperature and the environmental pH affect the potency of nanoparticles in the bioremediation process ( Tan et al., 2018 ). For instance, carbon dots nanoparticles have recently gained attention in the remediation of environmental pollutants owing to their abundance, low toxicity and unique optical properties ( Long et al., 2021 ). It is therefore necessary to carry out further research to unravel technologies and mechanisms to improve the efficiency of the bioremediation process.
10 Future perspectives and conclusions
A number of research endeavours have been carried out on the use of microbial enzymes for bioremediation of waste materials; however, it is very important to improve the process to ensure a safer and more sustainable environment. It is imperative to intensify research to unravel novel microbes that can effectively and rapidly bioremediate different pollutants, especially from industrial sources. Perhaps the novel microbes and their enzymes may have the inherent ability to bioremediate pollutants better than the presently used ones. It is also very important to carry out more studies to innovate rapid detection methods to reveal the progress or help to confirm total biodegradation of pollutants in the environment. Similarly, microbes presently used in bioremediation can be genetically modified to produce more enzymes which will enhance their biodegrading ability. A combination of different microbial consortium other than a single microbial consortium would be a better approach to bioremediation, as this would bring about the presence of different organisms which utilizes different substrate, consequently increasing the rate of microbial biodegradation.
Often, microbes are majorly used to degrade organic substrates, leaving out the persistent inorganic pollutants. Hence, research should be intensified to discover microbes that are capable of degrading inorganic (synthetic) pollutants. In recent years, nuclear wastes generated from the research sectors, hospitals, fuel processing plants and nuclear reactors have remained a global source of pollution. Therefore, the use of microbes and microbial enzymes in the bioremediation of nuclear wastes should be seriously taken into consideration. Occasionally, microbes themselves serve as a source of pollution instead of remediating pollutants. An example of such can be found when microbial biostimulation which results in algal bloom is carried out Consequently, methods to prevent this should be devised to ensure a sustainable environment.
Furthermore, in nature (outside the laboratory), the degradation of different compounds occurs at a different temperature, while the survival of microbes in nature are also environment-specific (temperature). It is therefore essential to carry out more field research to determine the optimum temperature for the degradation of different compounds in nature. In addition, it is also essential to find a balance between the environmental temperature and the temperature for the survival of different microbes in the environment. This would help to prevent bioremediation failure when external microbes are to be recruited or introduced to an environment. As positive and effective microbes might be recruited in the bioremediation of pollutants, it is important to carry out follow-up research to understand their effects on the environment after bioremediation, as some organisms which are introduced to an environment might later constitute pollution to the environment through mutation and other means. Hence, there should be regulatory bodies which would monitor the potential risk associated with microbes in specific environments.
Finally, if enzymes or microbes are directly applied to the soil, they might die or lose their potency before the remediation process begins; therefore, their combination with other agents, such as the nanoparticle could enhance their activity. More awareness is needed on the adoption of microbial degradation, and this will help policymakers as well as the populace to utilize this method. Many people unaware of this procedure might use the available conventional method, which might not be as safe and effective as the microbial biodegradation.
Author contributions
MA and OB conceptualized, wrote, reviewed, and edited the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the National Research Foundation, South Africa, grant number UID: 123634; 132595.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agrawal N., Verma P., Shahi S. K. (2018). Degradation of polycyclic aromatic hydrocarbons (phenanthrene and pyrene) by the ligninolytic fungi ganoderma lucidum isolated from the hardwood stump. Bioresour. Bioprocess. 5 (1), 1–9. doi: 10.1186/s40643-018-0197-5
CrossRef Full Text | Google Scholar
Albelda-Berenguer M., Monachon M., Joseph E. (2019). Siderophores: from natural roles to potential applications. Adv. Appl. Microbiol. 106, 193–225. doi: 10.1016/bs.aambs.2018.12.001
PubMed Abstract | CrossRef Full Text | Google Scholar
Al-Hawash A. B., Zhang J., Li S., Liu J., Ghalib H. B., Zhang X., et al. (2018). Biodegradation of n-hexadecane by aspergillus sp. RFC-1 and its mechanism. Ecotoxicol. Environ. Saf. 164, 398–408. doi: 10.1016/j.ecoenv.2018.08.049
AL-Huqail A. A., Kumar P., Eid E. M., Adelodun B., Abou Fayssal S., Singh J., et al. (2022). Risk assessment of heavy metals contamination in soil and two rice (Oryza sativa l.) varieties irrigated with paper mill effluent. Agriculture 12 (11), 1864.
Google Scholar
Alvarez L. M., Ruberto L. A. M., Gurevich J., Mac Cormack W. P. (2020). Environmental factors affecting reproducibility of bioremediation field assays in Antarctica. Cold Regions Sci. Technol. 169, 102915. doi: 10.1016/j.coldregions.2019.102915
Ambaye T. G., Vaccari M., Franzetti A., Prasad S., Formicola F., Rosatelli A., et al. (2023). Microbial electrochemical bioremediation of petroleum hydrocarbons (PHCs) pollution: recent advances and outlook. Chem. Eng. J. 452, 139372. doi: 10.1016/j.cej.2022.139372
Andreasen R., Li Y., Rehman Y., Ahmed M., Meyer R., Sabri A. (2018). Prospective role of indigenous Exiguobacterium profundum PT2 in arsenic biotransformation and biosorption by planktonic cultures and biofilms. J. Appl. Microbiol. 124 (2), 431–443.
PubMed Abstract | Google Scholar
Aragao W. A. B., Teixeira F. B., Fagundes N. C. F., Fernandes R. M., Fernandes L. M. P., da Silva M. C. F., et al. (2018). Hippocampal dysfunction provoked by mercury chloride exposure: evaluation of cognitive impairment, oxidative stress, tissue injury and nature of cell death. Oxid. Med. Cell. Longevity 2018 (1):1–11. doi: 10.1155/2018/7878050
Aregbesola O. A., Kumar A., Mokoena M. P., Olaniran A. O. (2020). Cloning, overexpression, purification, characterization and structural modelling of a metabolically active Fe2+ dependent 2, 6-dichloro-p-hydroquinone 1, 2-dioxygenase (CpsA) from bacillus cereus strain AOA-CPS_1. Int. J. Biol. Macromol. 161, 247–257. doi: 10.1016/j.ijbiomac.2020.05.268
Arstol E., Hohmann-Marriott M. F. (2019). Cyanobacterial siderophores–physiology, structure, biosynthesis, and applications. Mar. Drugs 17 (5), 281.
Atakpa E. O., Zhou H., Jiang L., Ma Y., Liang Y., Li Y., et al. (2022). Improved degradation of petroleum hydrocarbons by co-culture of fungi and biosurfactant-producing bacteria. Chemosphere 290, 133337. doi: 10.1016/j.chemosphere.2021.133337
Ayangbenro A. S., Babalola O. O. (2017). A new strategy for heavy metal polluted environments: a review of microbial biosorbents. Int. J. Environ. Res. Public Health 14 (1), 94. doi: 10.3390/ijerph14010094
Ayangbenro A. S., Babalola O. O. (2018). Metal(loid) bioremediation: strategies employed by microbial polymers. Sustainability 10 (9), 3028. doi: 10.3390/su10093028
Ayangbenro A. S., Babalola O. O. (2020). Genomic analysis of Bacillus cereus NWUAB01 and its heavy metal removal from polluted soil. Sci. Rep. 10 (1), 1–12. doi: 10.1038/s41598-020-75170-x
Ayangbenro A. S., Babalola O. O., Aremu O. S. (2019). Bioflocculant production and heavy metal sorption by metal resistant bacterial isolates from gold mining soil. Chemosphere 231, 113–120. doi: 10.1016/j.chemosphere.2019.05.092
Ayangbenro A. S., Olanrewaju O. S., Babalola O. O. (2018). Sulfate-reducing bacteria as an effective tool for sustainable acid mine bioremediation. Front. Microbiol. 1, 1986. doi: 10.3389/fmicb.2018.01986
Ayilara M. S., Olanrewaju O. S., Babalola O. O., Odeyemi O. (2020). Waste management through composting: challenges and potentials. Sustainability 12 (11), 4456. doi: 10.3390/su12114456
Babalola O. O., Aremu B. R., Ayangbenro A. S. (2019). Draft genome sequence of heavy metal-resistant bacillus cereus NWUAB01. Microbiol. Resour. Announc. 8 (7), e01706–e01718. doi: 10.1128/MRA.01706-18
Bai L., Zhang Q., Wang C., Yao X., Zhang H., Jiang H. (2019). Effects of natural dissolved organic matter on the complexation and biodegradation of 17α-ethinylestradiol in freshwater lakes. Environ. pollut. 246, 782–789. doi: 10.1016/j.envpol.2018.12.098
Bala S., Garg D., Thirumalesh B. V., Sharma M., Sridhar K., Inbaraj B. S., et al. (2022). Recent strategies for bioremediation of emerging pollutants: a review for a green and sustainable environment. Toxics 10 (8), 484. doi: 10.3390/toxics10080484
Bala G.-P., Rajnoveanu R.-M., Tudorache E., Motisan R., Oancea C. (2021). Air pollution exposure–the (in) visible risk factor for respiratory diseases. Environ. Sci. pollut. Res. 28, 19615–19628. doi: 10.1007/s11356-021-13208-x
Balali-Mood M., Naseri K., Tahergorabi Z., Khazdair M. R., Sadeghi M. (2021). Toxic mechanisms of five heavy metals: mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 12, 227. doi: 10.3389/fphar.2021.643972
Baltazar M., Gracioso L. H., Avanzi I. R., Karolski B., Tenório J. A. S., do Nascimento C. A. O., et al. (2019). Copper biosorption by rhodococcus erythropolis isolated from the sossego mine–PA–Brazil. J. Mater. Res. Technol. 8 (1), 475–483. doi: 10.1016/j.jmrt.2018.04.006
Bargiela R., Herbst F. A., Martínez-Martínez M., Seifert J., Rojo D., Cappello S., et al. (2015). Metaproteomics and metabolomics analyses of chronically petroleum-polluted sites reveal the importance of general anaerobic processes uncoupled with degradation. Proteomics 15 (20), 3508–3520. doi: 10.1002/pmic.201400614
Basu S., Rabara R. C., Negi S., Shukla P. (2018). Engineering PGPMOs through gene editing and systems biology: a solution for phytoremediation? Trends Biotechnol. 36 (5), 499–510.
Bhakat K., Chakraborty A., Islam E. (2019). Characterization of arsenic oxidation and uranium bioremediation potential of arsenic resistant bacteria isolated from uranium ore. Environ. Sci. pollut. Res. 26 (13), 12907–12919. doi: 10.1007/s11356-019-04827-6
Bhandari G., Gupta S., Chaudhary P., Chaudhary S., Gangola S. (2023). “Bioleaching: a sustainable resource recovery strategy for urban mining of e-waste,” in Microbial technology for sustainable e-waste management . Eds. Debbarma P., Kumar S., Suyal D. C., Soni R. (Cham: Springer), 157–175.
Bhandari S., Poudel D. K., Marahatha R., Dawadi S., Khadayat K., Phuyal S., et al. (2021). Microbial enzymes used in bioremediation. J. Chem. 2021, 1–17. doi: 10.1155/2021/8849512
Bharagava R. N., Purchase D., Saxena G., Mulla S. I. (2019). “Applications of metagenomics in microbial bioremediation of pollutants: from genomics to environmental cleanup,” in Microbial diversity in the genomic era (Amsterdam: Elsevier), 459–477.
Bhatt P., Gangola S., Joshi C., Chaudhary P., Kumar G., Bhandari G., et al. (2021a). Recent advancements and mechanism of microbial enzymes in sustainable agriculture. Microb. Technol. Sustain. Environ. 1, 247–259. doi: 10.1007/978-981-16-3840-4_15
Bhatt P., Verma A., Gangola S., Bhandari G., Chen S. (2021b). Microbial glycoconjugates in organic pollutant bioremediation: recent advances and applications. Microb. Cell Factories 20 (1), 1–18. doi: 10.1186/s12934-021-01556-9
Brinza L., Geraki K., Cojocaru C., Holdt S. L., Neamtu M. (2020). Baltic Fucus vesiculosus as potential bio-sorbent for zn removal: mechanism insight. Chemosphere 238, 124652. doi: 10.1016/j.chemosphere.2019.124652
Cao H.-L., Liu C., Cai F.-Y., Qiao X.-X., Dichiara A. B., Tian C., et al. (2020). In situ immobilization of ultra-fine Ag NPs onto magnetic Ag@RF@Fe3O4 core-satellite nanocomposites for the rapid catalytic reduction of nitrophenols. Water Res. 179, 115882. doi: 10.1016/j.watres.2020.115882
Chaudhary P., Ahamad L., Chaudhary A., Kumar G., Chen W.-J., Chen S. (2023a). Nanoparticle-mediated bioremediation as a powerful weapon in the removal of environmental pollutants. J. Environ. Chem. Eng. 11, 109591. doi: 10.1016/j.jece.2023.109591
Chaudhary P., Xu M., Ahamad L., Chaudhary A., Kumar G., Adeleke B. S., et al. (2023b). Application of synthetic consortia for improvement of soil fertility, pollution remediation, and agricultural productivity: a review. Agronomy 13 (3), 643. doi: 10.3390/agronomy13030643
Chellaiah E. (2018). Cadmium (heavy metals) bioremediation by Pseudomonas aeruginosa : a minireview. Appl. Water Sci. 8, 1–10. doi: 10.1007/s13201-018-0796-5
Chugh M., Kumar L., Shah M. P., Bharadvaja N. (2022). Algal bioremediation of heavy metals: an insight into removal mechanisms, recovery of by-products, challenges, and future opportunities. Energy Nexus 7, 100129. doi: 10.1016/j.nexus.2022.100129
Copete-Pertuz L. S., Plácido J., Serna-Galvis E. A., Torres-Palma R. A., Mora A. (2018). Elimination of isoxazolyl-penicillins antibiotics in waters by the ligninolytic native Colombian strain leptosphaerulina sp. considerations on biodegradation process and antimicrobial activity removal. Sci. Total Environ. 630, 1195–1204. doi: 10.1016/j.scitotenv.2018.02.244
Dave S., Das J. (2021). Role of microbial enzymes for biodegradation and bioremediation of environmental pollutants: challenges and future prospects. Bioremediat. Environ. Sustainability , 325–346. doi: 10.1016/B978-0-12-820524-2.00013-4
Dell’Anno F., Rastelli E., Tangherlini M., Corinaldesi C., Sansone C., Brunet C., et al. (2021). Highly contaminated marine sediments can host rare bacterial taxa potentially useful for bioremediation. Front. Microbiol. 12, 584850. doi: 10.3389/fmicb.2021.584850
Deng J., Fu D., Hu W., Lu X., Wu Y., Bryan H. (2020). Physiological responses and accumulation ability of microcystis aeruginosa to zinc and cadmium: implications for bioremediation of heavy metal pollution. Bioresour. Technol. 303, 122963. doi: 10.1016/j.biortech.2020.122963
Deng Y., Wang M., Tian T., Lin S., Xu P., Zhou L., et al. (2019). The effect of hexavalent chromium on the incidence and mortality of human cancers: a meta-analysis based on published epidemiological cohort studies. Front. Oncol. 9, 24. doi: 10.3389/fonc.2019.00024
de Souza M. B., de Souza Santos L. R., Borges R. E., Nunes H. F., Vieira T. B., Pacheco S. M., et al. (2020). Current status of ecotoxicological studies of bats in Brazil. Bull. Environ. Contam. Toxicol. 104, 393–399. doi: 10.1007/s00128-020-02794-0
Dhaka A., Chattopadhyay P. (2021). A review on physical remediation techniques for treatment of marine oil spills. J. Environ. Manage. 288, 112428. doi: 10.1016/j.jenvman.2021.112428
Duc H., Hung N., Oanh N. (2021). Anaerobic degradation of endosulfans by a mixed culture of pseudomonas sp. and staphylococcus sp. Appl. Biochem. Microbiol. 57 (3), 327–334. doi: 10.1134/S0003683821030030
Dusengemungu L., Kasali G., Gwanama C., Ouma K. O. (2020). Recent advances in biosorption of copper and cobalt by filamentous fungi. Front. Microbiol. 11, 3285. doi: 10.3389/fmicb.2020.582016
Dutta N., Usman M., Ashraf M. A., Luo G., Zhang S. (2022). Efficacy of emerging technologies in addressing reductive dechlorination for environmental bioremediation: a review. J. Hazard. Mater. Lett. 3, 100065. doi: 10.1016/j.hazl.2022.100065
Eisenhofer R., Minich J. J., Marotz C., Cooper A., Knight R., Weyrich L. S. (2019). Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 27 (2), 105–117. doi: 10.1016/j.tim.2018.11.003
Fashola M. O., Ngole-Jeme V. M., Babalola O. O. (2016). Heavy metal pollution from gold mines: environmental effects and bacterial strategies for resistance. Int. J. Environ. Res. Public Health 13 (11), 1047. doi: 10.3390/ijerph13111047
Fashola M. O., Ngole-Jeme V. M., Babalola O. O. (2020a). Heavy metal immobilization potential of indigenous bacteria isolated from gold mine tailings. Int. J. Environ. Res. 14 (1), 71–86. doi: 10.1007/s41742-019-00240-6
Fashola M. O., Ngole-Jeme V. M., Babalola O. O. (2020b). Physicochemical properties, heavy metals, and metal-tolerant bacteria profiles of abandoned gold mine tailings in krugersdorp, south Africa. Can. J. Soil Sci. 100 (3), 217–233. doi: 10.1139/cjss-2018-0161
Fay M. J., Alt L. A., Ryba D., Salamah R., Peach R., Papaeliou A., et al. (2018). Cadmium nephrotoxicity is associated with altered microRNA expression in the rat renal cortex. Toxics 6 (1), 16. doi: 10.3390/toxics6010016
Fu W., Xu M., Sun K., Hu L., Cao W., Dai C., et al. (2018). Biodegradation of phenanthrene by endophytic fungus phomopsis liquidambari in vitro and in vivo. Chemosphere 203, 160–169. doi: 10.1016/j.chemosphere.2018.03.164
Gangola S., Sharma A., Bhatt P., Khati P., Chaudhary P. (2018). Presence of esterase and laccase in bacillus subtilis facilitates biodegradation and detoxification of cypermethrin. Sci. Rep. 8 (1), 12755.
Gaur N., Narasimhulu K., PydiSetty Y. (2018). Recent advances in the bio-remediation of persistent organic pollutants and its effect on environment. J. Clean. Prod. 198, 1602–1631. doi: 10.1016/j.jclepro.2018.07.076
Gaur V. K., Tripathi V., Manickam N. (2022). “Bacterial-and fungal-mediated biodegradation of petroleum hydrocarbons in soil,” in Development in wastewater treatment research and processes (Amsterdam: Elsevier), 407–427.
Geetha N., Bhavya G., Abhijith P., Shekhar R., Dayananda K., Jogaiah S. (2021). Insights into nanomycoremediation: secretomics and mycogenic biopolymer nanocomposites for heavy metal detoxification. J. Hazard. Mater. 409, 124541. doi: 10.1016/j.jhazmat.2020.124541
Goswami M., Chakraborty P., Mukherjee K., Mitra G., Bhattacharyya P., Dey S., et al. (2018). Bioaugmentation and biostimulation: a potential strategy for environmental remediation. J. Microbiol. Exp. 6 (5), 223–231.
Gricajeva A., Nadda A. K., Gudiukaite R. (2022). Insights into polyester plastic biodegradation by carboxyl ester hydrolases. J. Chem. Technol. Biotechnol. 97 (2), 359–380. doi: 10.1002/jctb.6745
Hitt L. G., Khalil S., Blanchette A., Finkelstein M. E., Iverson E. N., McClelland S. C., et al. (2023). Lead exposure is correlated with reduced nesting success of an urban songbird. Environ. Res. 227 (1), 115711. doi: 10.1016/j.envres.2023.115711
Huang Y., Pan H., Wang Q., Ge Y., Liu W., Christie P. (2019). Enrichment of the soil microbial community in the bioremediation of a petroleum-contaminated soil amended with rice straw or sawdust. Chemosphere 224, 265–271. doi: 10.1016/j.chemosphere.2019.02.148
Hussain A., Rehman F., Rafeeq H., Waqas M., Asghar A., Afsheen N., et al. (2022). In-situ, ex-situ, and nano-remediation strategies to treat polluted soil, water, and air–a review. Chemosphere 289, 133252. doi: 10.1016/j.chemosphere.2021.133252
Iark D., dos Reis Buzzo A. J., Garcia J. A. A., Côrrea V. G., Helm C. V., Corrêa R. C. G., et al. (2019). Enzymatic degradation and detoxification of azo dye Congo red by a new laccase from Oudemansiella canarii . Bioresour. Technol. 289, 121655. doi: 10.1016/j.biortech.2019.121655
Imam A., Suman S. K., Ghosh D., Kanaujia P. K. (2019). Analytical approaches used in monitoring the bioremediation of hydrocarbons in petroleum-contaminated soil and sludge. TrAC Trends Anal. Chem. 118, 50–64. doi: 10.1016/j.trac.2019.05.023
Imron M. F., Kurniawan S. B., Ismail N. I., Abdullah S. R. S. (2020). Future challenges in diesel biodegradation by bacteria isolates: a review. J. Clean. Prod. 251, 119716. doi: 10.1016/j.jclepro.2019.119716
Jacob J. M., Karthik C., Saratale R. G., Kumar S. S., Prabakar D., Kadirvelu K., et al. (2018). Biological approaches to tackle heavy metal pollution: a survey of literature. J. Environ. Manage. 217, 56–70. doi: 10.1016/j.jenvman.2018.03.077
Jafari A., Ghaderpoori M., Kamarehi B., Abdipour H. (2019). Soil pollution evaluation and health risk assessment of heavy metals around douroud cement factory, Iran. Environ. Earth Sci. 78 (8), 1–9. doi: 10.1007/s12665-019-8220-5
Kebede G., Tafese T., Abda E. M., Kamaraj M., Assefa F. (2021). Factors influencing the bacterial bioremediation of hydrocarbon contaminants in the soil: mechanisms and impacts. J. Chem. 2021, 1–17. doi: 10.1155/2021/9823362
Khan A., Singh P., Srivastava A. (2018). Synthesis, nature and utility of universal iron chelator–siderophore: a review. Microbiol. Res. 212, 103–111. doi: 10.1016/j.micres.2017.10.012
Kharangate-Lad A., D’Souza N. C. (2021). Current approaches in bioremediation of toxic contaminants by application of microbial cells; biosurfactants and bioemulsifiers of microbial origin. Rhizobiont Bioremediat. Hazard. Waste , 217–263. doi: 10.1007/978-981-16-0602-1_11
Kour D., Khan S. S., Kour H., Kaur T., Devi R., Rai P. K., et al. (2022). Microbe-mediated bioremediation: current research and future challenges. J. Appl. Biol. Biotechnol. 10 (2), 6–24. doi: 10.7324/JABB.2022.10s202
Kumar V., Garg V. K., Kumar S., Biswas J. K. (2022). Omics for environmental engineering and microbiology systems (Florida: CRC Press).
Kumar G., Lal S., Maurya S. K., Bhattacherjee A., Chaudhary P., Gangola S., et al. (2021). Exploration of klebsiella pneumoniae M6 for paclobutrazol degradation, plant growth attributes, and biocontrol action under subtropical ecosystem. PloS One 16 (12), e0261338. doi: 10.1371/journal.pone.0261338
Kumar G., Lal S., Soni S. K., Maurya S. K., Shukla P. K., Chaudhary P., et al. (2022). Mechanism and kinetics of chlorpyrifos co-metabolism by using environment restoring microbes isolated from rhizosphere of horticultural crops under subtropics. Front. Microbiol. 13, 2796. doi: 10.3389/fmicb.2022.891870
Kumar A., Sharma A., Chaudhary P., Gangola S. (2021). Chlorpyrifos degradation using binary fungal strains isolated from industrial waste soil. Biologia 76 (10), 3071–3080. doi: 10.1007/s11756-021-00816-8
Kumari A., Kaur R., Kaur R. (2019). A review on fate and remediation techniques of oil spills. Int. J. Res. Pharm. Sci. 10, 111.
Kumari V., Tripathi A. (2020). Remediation of heavy metals in pharmaceutical effluent with the help of bacillus cereus-based green-synthesized silver nanoparticles supported on alumina. Appl. Nanosci. 10 (6), 1709–1719. doi: 10.1007/s13204-020-01351-9
Kushwaha A., Maurya S., Pathak R. K., Agarwal S., Chaurasia P. K., Singh M. (2018). “Laccase from white rot fungi having significant role in food, pharma, and other industries,” in Research advancements in pharmaceutical, nutritional, and industrial enzymology (Pennsylvania: IGI Global), 253–277.
Li Q., Liu J., Gadd G. M. (2020). Fungal bioremediation of soil co-contaminated with petroleum hydrocarbons and toxic metals. Appl. Microbiol. Biotechnol. 104 (21), 8999–9008. doi: 10.1007/s00253-020-10854-y
Li N., Xia Q., Niu M., Ping Q., Xiao H. (2018). Immobilizing laccase on different species wood biochar to remove the chlorinated biphenyl in wastewater. Sci. Rep. 8 (1), 1–9. doi: 10.1038/s41598-018-32013-0
Li F., Zheng Y., Tian J., Ge F., Liu X., Tang Y., et al. (2019). Cupriavidus sp. strain Cd02-mediated pH increase favoring bioprecipitation of Cd2+ in medium and reduction of cadmium bioavailability in paddy soil. Ecotoxicol. Environ. Saf. 184, 109655.
Liu Z., Shao B., Zeng G., Chen M., Li Z., Liu Y., et al. (2018). Effects of rhamnolipids on the removal of 2, 4, 2, 4-tetrabrominated biphenyl ether (BDE-47) by phanerochaete chrysosporium analyzed with a combined approach of experiments and molecular docking. Chemosphere 210, 922–930. doi: 10.1016/j.chemosphere.2018.07.114
Liu X., Shi H., Bai Z., Zhou W., Liu K., Wang M., et al. (2020). Heavy metal concentrations of soils near the large opencast coal mine pits in China. Chemosphere 244, 125360. doi: 10.1016/j.chemosphere.2019.125360
Liu L., Wu Q., Miao X., Fan T., Meng Z., Chen X., et al. (2022). Study on toxicity effects of environmental pollutants based on metabolomics: a review. Chemosphere 286, 131815. doi: 10.1016/j.chemosphere.2021.131815
Long C., Jiang Z., Shangguan J., Qing T., Zhang P., Feng B. (2021). Applications of carbon dots in environmental pollution control: a review. Chem. Eng. J. 406, 126848. doi: 10.1016/j.cej.2020.126848
Lu J., Zhang C., Leong H. Y., Show P. L., Lu F., Lu Z. (2020). Overproduction of lipoxygenase from pseudomonas aeruginosa in Escherichia coli by auto-induction expression and its application in triphenylmethane dyes degradation. J. Biosci. Bioeng. 129 (3), 327–332. doi: 10.1016/j.jbiosc.2019.09.006
Luo H., Yuan D. B., Zhang W. (2015). Association between cadmium exposure and renal cancer risk: a meta-analysis of observational studies. Sci. Rep. 5 (1), 1–8.
Machado L. F., de Assis Leite D. C., da Costa Rachid C. T. C., Paes J. E., Martins E. F., Peixoto R. S., et al. (2019). Tracking mangrove oil bioremediation approaches and bacterial diversity at different depths in an in situ mesocosms system. Front. Microbiol. 10, 2107. doi: 10.3389/fmicb.2019.02107
Mahmood A., Bilal B., Naeem Z., Iram S. (2021). “Physical, chemical, and biological remediation techniques for textile effluents in context with developed and developing countries,” in Rhizobiont in bioremediation of hazardous waste (Singapore: Springer), 409–441.
Mahmoud G. A.-E. (2021). “Microbial scavenging of heavy metals using bioremediation strategies,” in Rhizobiont in bioremediation of hazardous waste . Eds. Vivek K., Ram P., Manoj K. (Singapore: Springer), 265–289.
Maity J. P., Chen G.-S., Huang Y.-H., Sun A.-C., Chen C.-Y. (2019). Ecofriendly heavy metal stabilization: microbial induced mineral precipitation (MIMP) and biomineralization for heavy metals within the contaminated soil by indigenous bacteria. Geomicrobiol. J. 36 (7), 612–623. doi: 10.1080/01490451.2019.1597216
Malakar N., Mitra S., Toppo P., Mathur P. (2020). Understanding the functional attributes of different microbial enzymes in bioremediation. NBU J. Plant Sci. 12 (1), 58–69. doi: 10.55734/NBUJPS.2020.v12i01.005
Manisalidis I., Stavropoulou E., Stavropoulos A., Bezirtzoglou E. (2020). Environmental and health impacts of air pollution: a review. Front. Public Health 1), 14. doi: 10.3389/fpubh.2020.00014
Mathew B. B., Krishnamurthy N. B. (2018). Screening and identification of bacteria isolated from industrial area groundwater to study lead sorption: kinetics and statistical optimization of biosorption parameters. Groundwater Sustain. Dev. 7, 313–327. doi: 10.1016/j.gsd.2018.07.007
Medfu Tarekegn M., Zewdu Salilih F., Ishetu A. I. (2020). Microbes used as a tool for bioremediation of heavy metal from the environment. Cogent Food Agric. 6 (1), 1783174. doi: 10.1080/23311932.2020.1783174
Methneni N., Morales-Gonzalez J. A., Jaziri A., Mansour H. B., Fernandez-Serrano M. (2021). Persistent organic and inorganic pollutants in the effluents from the textile dyeing industries: ecotoxicology appraisal via a battery of biotests. Environ. Res. 196, 110956. doi: 10.1016/j.envres.2021.110956
Miri S., Rasooli A., Brar S. K., Rouissi T., Martel R. (2022). Biodegradation of p-xylene–a comparison of three psychrophilic pseudomonas strains through the lens of gene expression. Environ. Sci. pollut. Res. 29 (15), 21465–21479. doi: 10.1007/s11356-021-17387-5
Mir-Tutusaus J. A., Baccar R., Caminal G., Sarrà M. (2018). Can white-rot fungi be a real wastewater treatment alternative for organic micropollutants removal? a review. Water Res. 138, 137–151. doi: 10.1016/j.watres.2018.02.056
Mohamed M. S., El-Arabi N. I., El-Hussein A., El-Maaty S. A., Abdelhadi A. A. (2020). Reduction of chromium-VI by chromium-resistant escherichia coli FACU: a prospective bacterium for bioremediation. Folia Microbiol. 65 (4), 687–696. doi: 10.1007/s12223-020-00771-y
Mohapatra B., Phale P. S. (2021). Microbial degradation of naphthalene and substituted naphthalenes: metabolic diversity and genomic insight for bioremediation. Front. Bioeng. Biotechnol. 9, 602445. doi: 10.3389/fbioe.2021.602445
Mohd S., Kushwaha A. S., Shukla J., Mandrah K., Shankar J., Arjaria N., et al. (2019). Fungal mediated biotransformation reduces toxicity of arsenic to soil dwelling microorganism and plant. Ecotoxicol. Environ. Saf. 176, 108–118. doi: 10.1016/j.ecoenv.2019.03.053
Moreira V., Lebron Y., Lange L., Santos L. (2019). Simultaneous biosorption of cd (II), Ni (II) and Pb (II) onto a brown macroalgae fucus vesiculosus: mono-and multi-component isotherms, kinetics and thermodynamics. J. Environ. Manage. 251, 109587. doi: 10.1016/j.jenvman.2019.109587
Mousavi S. M., Hashemi S. A., Iman Moezzi S. M., Ravan N., Gholami A., Lai C. W., et al. (2021). Recent advances in enzymes for the bioremediation of pollutants. Biochem. Res. Int. 2021, 1–12.
Mukherjee A., Reddy M. S. (2020). Metatranscriptomics: an approach for retrieving novel eukaryotic genes from polluted and related environments. 3 Biotech. 10 (2), 71. doi: 10.1007/s13205-020-2057-1
Mukjang N., Chitov T., Mhuantong W., Champreda V., Pathom-Aree W., Sattayawat P., et al. (2022). Bacterial communities associated with crude oil bioremediation through composting approaches with indigenous bacterial isolate. Life 12 (11), 1712. doi: 10.3390/life12111712
Nanda M., Kumar V., Sharma D. (2019). Multimetal tolerance mechanisms in bacteria: the resistance strategies acquired by bacteria that can be exploited to ‘clean-up’heavy metal contaminants from water. Aquat. Toxicol. 212, 1–10. doi: 10.1016/j.aquatox.2019.04.011
Narayanan M., Ali S. S., El-Sheekh M. (2023). A comprehensive review on the potential of microbial enzymes in multipollutant bioremediation: MechanAisms, challenges, and future prospects. J. Environ. Manage. 334, 117532. doi: 10.1016/j.jenvman.2023.117532
Nath S., Deb B., Sharma I. (2018). Isolation of toxic metal-tolerant bacteria from soil and examination of their bioaugmentation potentiality by pot studies in cadmium-and lead-contaminated soil. Int. Microbiol. 21, 35–45. doi: 10.1007/s10123-018-0003-4
Ndeddy Aka R. J., Babalola O. O. (2016). Effect of bacterial inoculation of strains of pseudomonas aeruginosa, alcaligenes feacalis and bacillus subtilis on germination, growth and heavy metal (Cd, cr, and Ni) uptake of brassica juncea. Int. J. Phytoremediation 18 (2), 200–209.
Ndeddy Aka R. J., Babalola O. O. (2017). Identification and characterization of cr-, cd-, and Ni-tolerant bacteria isolated from mine tailings. Bioremediat. J. 21 (1), 1–19.
Nivetha N., Srivarshine B., Sowmya B., Rajendiran M., Saravanan P., Rajeshkannan R., et al. (2022). A comprehensive review on bio-stimulation and bio-enhancement towards remediation of heavy metals degeneration. Chemosphere 3, 137099.
Nowicka B., Fesenko T., Walczak J., Kruk J. (2020). The inhibitor-evoked shortage of tocopherol and plastoquinol is compensated by other antioxidant mechanisms in chlamydomonas reinhardtii exposed to toxic concentrations of cadmium and chromium ions. Ecotoxicol. Environ. Saf. 191, 110241. doi: 10.1016/j.ecoenv.2020.110241
Ogunlaja A., Ogunlaja O. O., Okewole D. M., Morenikeji O. A. (2019). Risk assessment and source identification of heavy metal contamination by multivariate and hazard index analyses of a pipeline vandalised area in Lagos state, Nigeria. Sci. Total Environ. 651, 2943–2952. doi: 10.1016/j.scitotenv.2018.09.386
Ojuederie O. B., Babalola O. O. (2017). Microbial and plant-assisted bioremediation of heavy metal polluted environments: a review. Int. J. Environ. Res. Public Health 14 (12), 1504. doi: 10.3390/ijerph14121504
Ojuederie O. B., Chukwuneme F., Olanrewaju O., Ayilara M., Adegboyega T. T., Babalola O. O. (2021). Contribution of microbial inoculants in sustainable maintenance of human health, including test methods and evaluation of safety of microbial pesticide microorganisms. Biopesticides: Botanicals Microorganisms Improving Agric. Hum. Health 2021, 207–240.
Orlovic-Leko P., Farkas B., Galic I. (2022). A short review of environmental and health impacts of gold mining. Reliability: Theory Appl. 17(SI 4 (SI 4 (70), 242–248.
Pande V., Pandey S. C., Sati D., Pande V., Samant M. (2020). Bioremediation: an emerging effective approach towards environment restoration. Environ. Sustainability 3 (1), 91–103. doi: 10.1007/s42398-020-00099-w
Patel A. B., Jain K. R., Manvar T., Desai C., Madamwar D. (2022). Enriched bacterial community efficiently degrade polycyclic aromatic hydrocarbons in soil ecosystem: insights from a mesocosms study. Biochem. Eng. J. 185, 108516. doi: 10.1016/j.bej.2022.108516
Patel A. K., Singhania R. R., Albarico F. P. J. B., Pandey A., Chen C.-W., Dong C.-D. (2022). Organic wastes bioremediation and its changing prospects. Sci. Total Environ. 824, 153889. doi: 10.1016/j.scitotenv.2022.153889
Pavesi T., Moreira J. C. (2020). Mechanisms and individuality in chromium toxicity in humans. J. Appl. Toxicol. 40 (9), 1183–1197. doi: 10.1002/jat.3965
Perera I. C., Hemamali E. H. (2022). Genetically modified organisms for bioremediation: current research and advancements. Bioremediat. Environ. Pollut.: Emerg. Trends Strategies 1, 163–186.
Pope C. A., Lefler J. S., Ezzati M., Higbee J. D., Marshall J. D., Kim S.-Y., et al. (2019). Mortality risk and fine particulate air pollution in a large, representative cohort of US adults. Environ. Health Perspect. 127 (7), 077007.
Prabagar S., Dharmadasa R. M., Lintha A., Thuraisingam S., Prabagar J. (2021). Accumulation of heavy metals in grape fruit, leaves, soil and water: a study of influential factors and evaluating ecological risks in jaffna, Sri Lanka. Environ. Sustainability Indic. 12, 100147. doi: 10.1016/j.indic.2021.100147
Pratush A., Kumar A., Hu Z. (2018). Adverse effect of heavy metals (As, Pb, Hg, and cr) on health and their bioremediation strategies: a review. Int. Microbiol. 21 (3), 97–106. doi: 10.1007/s10123-018-0012-3
Qiao W., Zhang Y., Xia H., Luo Y., Liu S., Wang S., et al. (2019). Bioimmobilization of lead by bacillus subtilis X3 biomass isolated from lead mine soil under promotion of multiple adsorption mechanisms. R. Soc. Open Sci. 6 (2), 181701. doi: 10.1098/rsos.181701
Rabani M. S., Sharma R., Singh R., Gupta M. (2022). Characterization and identification of naphthalene degrading bacteria isolated from petroleum contaminated sites and their possible use in bioremediation. Polycyclic Aromatic Compounds 42 (3), 978–989. doi: 10.1080/10406638.2020.1759663
Rai P. K., Lee S. S., Zhang M., Tsang Y. F., Kim K.-H. (2019). Heavy metals in food crops: health risks, fate, mechanisms, and management. Environ. Int. 125 (1), 365–385. doi: 10.1016/j.envint.2019.01.067
Rehman A. U., Nazir S., Irshad R., Tahir K., ur Rehman K., Islam R. U., et al. (2021). Toxicity of heavy metals in plants and animals and their uptake by magnetic iron oxide nanoparticles. J. Mol. Liquids 321, 114455. doi: 10.1016/j.molliq.2020.114455
Ren X., Zeng G., Tang L., Wang J., Wan J., Wang J., et al. (2018). The potential impact on the biodegradation of organic pollutants from composting technology for soil remediation. Waste Manage. 72, 138–149. doi: 10.1016/j.wasman.2017.11.032
Saavedra R., Muñoz R., Taboada M. E., Vega M., Bolado S. (2018). Comparative uptake study of arsenic, boron, copper, manganese and zinc from water by different green microalgae. Bioresour. Technol. 263, 49–57. doi: 10.1016/j.biortech.2018.04.101
Saha L., Tiwari J., Bauddh K., Ma Y. (2021). Recent developments in microbe–plant-based bioremediation for tackling heavy metal-polluted soils. Front. Microbiol. 12, 731723. doi: 10.3389/fmicb.2021.731723
Sahay R. (2021). Synthetic applications of laccase and its catalytic potentials. Inter J. Adv. Eng. Res. Sci. 8 (6), 112–120. doi: 10.22161/ijaers.86.12
Sangkharak K., Choonut A., Rakkan T., Prasertsan P. (2020). The degradation of phenanthrene, pyrene, and fluoranthene and its conversion into medium-chain-length polyhydroxyalkanoate by novel polycyclic aromatic hydrocarbon-degrading bacteria. Curr. Microbiol. 77 (6), 897–909. doi: 10.1007/s00284-020-01883-x
Saranya P., Selvi P., Sekaran G. (2019). Integrated thermophilic enzyme-immobilized reactor and high-rate biological reactors for treatment of palm oil-containing wastewater without sludge production. Bioprocess Biosyst. Eng. 42, 1053–1064. doi: 10.1007/s00449-019-02104-x
Sarmah P., Rout J. (2020). “Role of algae and cyanobacteria in bioremediation: prospects in polyethylene biodegradation,” in Advances in cyanobacterial biology (Amsterdam: Elsevier), 333–349.
Saxena G., Kishor R., Bharagava R. N. (2020). “Application of microbial enzymes in degradation and detoxification of organic and inorganic pollutants,” in Bioremediation of industrial waste for environmental safety (New York: Springer), 41–51.
Sayed K., Baloo L., Sharma N. K. (2021). Bioremediation of total petroleum hydrocarbons (TPH) by bioaugmentation and biostimulation in water with floating oil spill containment booms as bioreactor basin. Int. J. Environ. Res. Public Health 18 (5), 2226. doi: 10.3390/ijerph18052226
Sedlakova-Kadukova J., Kopcakova A., Gresakova L., Godany A., Pristas P. (2019). Bioaccumulation and biosorption of zinc by a novel streptomyces K11 strain isolated from highly alkaline aluminium brown mud disposal site. Ecotoxicol. Environ. Saf. 167, 204–211. doi: 10.1016/j.ecoenv.2018.09.123
Sen S. K., Patra P., Das C. R., Raut S., Raut S. (2019). Pilot-scale evaluation of bio-decolorization and biodegradation of reactive textile wastewater: an impact on its use in irrigation of wheat crop. Water Resour. Industry 21, 100106. doi: 10.1016/j.wri.2019.100106
Shah H., Jain S. (2020). “Bioremediation: an approach for environmental pollutants detoxification,” in Waste to energy: prospects and applications (New York: Springer), 121–142.
Shanmuganathan R., Karuppusamy I., Saravanan M., Muthukumar H., Ponnuchamy K., Ramkumar V. S., et al. (2019). Synthesis of silver nanoparticles and their biomedical applications-a comprehensive review. Curr. Pharm. Design 25 (24), 2650–2660. doi: 10.2174/1381612825666190708185506
Sharma P., Dutta D., Udayan A., Nadda A. K., Lam S. S., Kumar S. (2022a). Role of microbes in bioaccumulation of heavy metals in municipal solid waste: impacts on plant and human being. Environ. pollut. 305, 119248. doi: 10.1016/j.envpol.2022.119248
Sharma M., Nandy A., Taylor N., Venkatesan S. V., Kollath V. O., Karan K., et al. (2020). Bioelectrochemical remediation of phenanthrene in a microbial fuel cell using an anaerobic consortium enriched from a hydrocarbon-contaminated site. J. Hazard. Mater. 389, 121845. doi: 10.1016/j.jhazmat.2019.121845
Sharma P., Singh S. P., Iqbal H. M., Tong Y. W. (2022b). Omics approaches in bioremediation of environmental contaminants: an integrated approach for environmental safety and sustainability. Environ. Res. 211, 113102. doi: 10.1016/j.envres.2022.113102
Sharma P., Singh S. P., Parakh S. K., Tong Y. W. (2022c). Health hazards of hexavalent chromium (Cr (VI)) and its microbial reduction. Bioengineered 13 (3), 4923–4938. doi: 10.1080/21655979.2022.2037273
Singh R. K., Tripathi R., Ranjan A., Srivastava A. K. (2020). “Fungi as potential candidates for bioremediation,” in Abatement of environmental pollutants (Amsterdam: Elsevier), 177–191.
Siric I., Eid E. M., El-Morsy M. H., Osman H. E., Adelodun B., Abou Fayssal S., et al. (2022a). Health risk assessment of hazardous heavy metals in two varieties of mango fruit (Mangifera indica l. var. dasheri and langra). Horticulturae 8 (9), 832. doi: 10.3390/horticulturae8090832
Siric I., Eid E. M., Taher M. A., El-Morsy M. H., Osman H. E., Kumar P., et al. (2022b). Combined use of spent mushroom substrate biochar and PGPR improves growth, yield, and biochemical response of cauliflower (Brassica oleracea var. botrytis): a preliminary study on greenhouse cultivation. Horticulturae 8 (9), 830. doi: 10.3390/horticulturae8090830
Sonawane J. M., Rai A. K., Sharma M., Tripathi M., Prasad R. (2022). Microbial biofilms: recent advances and progress in environmental bioremediation. Sci. Total Environ. 824, 153843. doi: 10.1016/j.scitotenv.2022.153843
Song J., Zhang S., Xie Y., Li Q. (2019). Purification and characteristics of an aflatoxin B1 degradation enzyme isolated from pseudomonas aeruginosa. FEMS Microbiol. Lett. 366 (5), fnz034. doi: 10.1093/femsle/fnz034
Sonune N. (2021). “Microbes: a potential tool for bioremediation,” in Rhizobiont in bioremediation of hazardous waste . Eds. Vivek K., Ram P., Manoj K. (Singapore: Springer), 391–407.
Sonune N., Garode A. (2018). Isolation, characterization and identification of extracellular enzyme producer bacillus licheniformis from municipal wastewater and evaluation of their biodegradability. Biotechnol. Res. Innovation 2 (1), 37–44. doi: 10.1016/j.biori.2018.03.001
Sravya K., Sangeetha S. (2022). Feasibility study on bioremediation techniques to contaminated soils. Mater. Today: Proc. 51, 2556–2560. doi: 10.1016/j.matpr.2021.12.364
Sun W., Cheng K., Sun K. Y., Ma X. (2021). Microbially mediated remediation of contaminated sediments by heavy metals: a critical review. Curr. pollut. Rep. 7 (2), 201–212. doi: 10.1007/s40726-021-00175-7
Sun L., Guo D., Liu K., Meng H., Zheng Y., Yuan F., et al. (2019). Levels, sources, and spatial distribution of heavy metals in soils from a typical coal industrial city of tangshan, China. Catena 175, 101–109. doi: 10.1016/j.catena.2018.12.014
Tak H. I., Ahmad F., Babalola O. O. (2012). Advances in the application of plant growth-promoting rhizobacteria in phytoremediation of heavy metals. Rev. Environ. Contam. Toxicol. 223 (1), 33–52.
Tan W., Peralta-Videa J. R., Gardea-Torresdey J. L. (2018). Interaction of titanium dioxide nanoparticles with soil components and plants: current knowledge and future research needs–a critical review. Environ. Sci.: Nano 5 (2), 257–278. doi: 10.1039/C7EN00985B
Taran M., Fateh R., Rezaei S., Gholi M. K. (2019). Isolation of arsenic accumulating bacteria from garbage leachates for possible application in bioremediation. Iranian J. Microbiol. 11 (1), 60. doi: 10.18502/ijm.v11i1.707
Thakare M., Sarma H., Datar S., Roy A., Pawar P., Gupta K., et al. (2021). Understanding the holistic approach to plant-microbe remediation technologies for removing heavy metals and radionuclides from soil. Curr. Res. Biotechnol. 3, 84–98. doi: 10.1016/j.crbiot.2021.02.004
Thakur M., Medintz I. L., Walper S. A. (2019). Enzymatic bioremediation of organophosphate compounds–progress and remaining challenges. Front. Bioeng. Biotechnol. 7, 289. doi: 10.3389/fbioe.2019.00289
Tian Q., Dou X., Huang L., Wang L., Meng D., Zhai L., et al. (2020). Characterization of a robust cold-adapted and thermostable laccase from pycnoporus sp. SYBC-L10 with a strong ability for the degradation of tetracycline and oxytetracycline by laccase-mediated oxidation. J. Hazard. Mater. 382, 121084.
Unuofin J. O., Falade A. O., Aladekoyi O. J. (2021). Applications of microbial laccases in bioremediation of environmental pollutants: potential issues, challenges, and prospects. Bioremediat. Environ. Sustainability 1, 519–540. doi: 10.1016/B978-0-12-820524-2.00021-3
Unuofin J. O., Okoh A. I., Nwodo U. U. (2019). Aptitude of oxidative enzymes for treatment of wastewater pollutants: a laccase perspective. Molecules 24 (11), 2064. doi: 10.3390/molecules24112064
Verma S., Kuila A. (2019). Bioremediation of heavy metals by microbial process. Environ. Technol. Innovation 14, 100369. doi: 10.1016/j.eti.2019.100369
Verma S., Verma P. K., Chakrabarty D. (2019). Arsenic bio-volatilization by engineered yeast promotes rice growth and reduces arsenic accumulation in grains. Int. J. Environ. Res. 13 (3), 475–485. doi: 10.1007/s41742-019-00188-7
Vo M.-T., Nguyen V.-T., Dao T.-S. (2020). Responses of green algae and diatom upon exposure to chromium and cadmium. Vietnam J. Sci. Technol. Eng. 62 (1), 69–73. doi: 10.31276/VJSTE.62(1).69-73
Vocciante M., Finocchi A., De Folly D′ Auris A., Conte A., Tonziello J., Pola A., et al. (2019). Enhanced oil spill remediation by adsorption with interlinked multilayered graphene. Materials 12 (14), 2231. doi: 10.3390/ma12142231
Wang L., Liu Y., Shu X., Lu S., Xie X., Shi Q. (2019). Complexation and conformation of lead ion with poly-γ-glutamic acid in soluble state. PloS One 14 (9), e0218742. doi: 10.1371/journal.pone.0218742
Wang Z., Luo Z., Yan C., Rosenfeldt R. R., Seitz F., Gui H. (2018). Biokinetics of arsenate accumulation and release in microcystis aeruginosa regulated by common environmental factors: practical implications for enhanced bioremediation. J. Clean. Prod. 199, 112–120. doi: 10.1016/j.jclepro.2018.07.131
Wang Y., Mandal A. K., Son Y.-O., Pratheeshkumar P., Wise J. T., Wang L., et al. (2018). Roles of ROS, Nrf2, and autophagy in cadmium-carcinogenesis and its prevention by sulforaphane. Toxicol. Appl. Pharmacol. 353, 23–30. doi: 10.1016/j.taap.2018.06.003
Wei Y., Fu J., Wu J., Jia X., Zhou Y., Li C., et al. (2018). Bioinformatics analysis and characterization of highly efficient polyvinyl alcohol (PVA)-degrading enzymes from the novel PVA degrader stenotrophomonas rhizophila QL-P4. Appl. Environ. Microbiol. 84 (1), e01898–e01817. doi: 10.1128/AEM.01898-17
Wu W., Jiang S., Zhao Q., Zhang K., Wei X., Zhou T., et al. (2018). Environmental exposure to metals and the risk of hypertension: a cross-sectional study in China. Environ. pollut. 233, 670–678. doi: 10.1016/j.envpol.2017.10.111
Wu P., Zhang Z., Luo Y., Bai Y., Fan J. (2022). Bioremediation of phenolic pollutants by algae-current status and challenges. Bioresour. Technol. 350, 126930. doi: 10.1016/j.biortech.2022.126930
Xiao F., Cheng J., Cao W., Yang C., Chen J., Luo Z. (2019). Removal of heavy metals from aqueous solution using chitosan-combined magnetic biochars. J. Colloid Interface Sci. 540, 579–584. doi: 10.1016/j.jcis.2019.01.068
Xie Q., Liu N., Lin D., Qu R., Zhou Q., Ge F. (2020). The complexation with proteins in extracellular polymeric substances alleviates the toxicity of cd (II) to chlorella vulgaris. Environ. pollut. 263, 114102. doi: 10.1016/j.envpol.2020.114102
Xu Q., Wu B., Chai X. (2022). In situ remediation technology for heavy metal contaminated sediment: a review. Int. J. Environ. Res. Public Health 19 (24), 16767. doi: 10.3390/ijerph192416767
Ye X., Peng T., Feng J., Yang Q., Pratush A., Xiong G., et al. (2019). A novel dehydrogenase 17β-HSDx from rhodococcus sp. P14 with potential application in bioremediation of steroids contaminated environment. J. Hazard. Mater. 362, 170–177. doi: 10.1016/j.jhazmat.2018.09.023
Zacharia J. T. (2019). Degradation pathways of persistent organic pollutants (POPs) in the environment. Persistent Organic Pollutants , 17–30. doi: 10.5772/intechopen.79645
Zeng F., Qiu B., Wu X., Niu S., Wu F., Zhang G. (2012). Glutathione-mediated alleviation of chromium toxicity in rice plants. Biol. Trace Elem. Res. 148, 255–263. doi: 10.1007/s12011-012-9362-4
Zhang C., Gan C., Ding L., Xiong M., Zhang A., Li P. (2020). Maternal inorganic mercury exposure and renal effects in the wanshan mercury mining area, southwest China. Ecotoxicol. Environ. Saf. 189, 109987. doi: 10.1016/j.ecoenv.2019.109987
Zhao B., Johnston F. H., Salimi F., Kurabayashi M., Negishi K. (2020). Short-term exposure to ambient fine particulate matter and out-of-hospital cardiac arrest: a nationwide case-crossover study in Japan. Lancet Planet. Health 4 (1), e15–e23. doi: 10.1016/S2542-5196(19)30262-1
Zhou Y., Liu X., Wang J. (2019). Characterization of microplastics and the association of heavy metals with microplastics in suburban soil of central China. Sci. Total Environ. 694, 133798. doi: 10.1016/j.scitotenv.2019.133798
Zuhara S., Isaifan R. (2018). The impact of criteria air pollutants on soil and water: a review. J. Environ. Sci. pollut. Res. 4 (2), 278–284. doi: 10.30799/jespr.133.18040205
Zwolak A., Sarzyńska M., Szpyrka E., Stawarczyk K. (2019). Sources of soil pollution by heavy metals and their accumulation in vegetables: a review. Water Air Soil pollut. 230 (7), 1–9. doi: 10.1007/s11270-019-4221-y
Keywords: microbial bioremediation, bioaugmentation, biostimulation, siderophores, biosorption
Citation: Ayilara MS and Babalola OO (2023) Bioremediation of environmental wastes: the role of microorganisms. Front. Agron. 5:1183691. doi: 10.3389/fagro.2023.1183691
Received: 10 March 2023; Accepted: 18 May 2023; Published: 30 May 2023.
Reviewed by:
Copyright © 2023 Ayilara and Babalola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olubukola O. Babalola, b2x1YnVrb2xhLmJhYmFsb2xhQG53dS5hYy56YQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.

IMAGES
COMMENTS
Bioremediation, an advanced and environmentally sustainable technology, utilizes biological microorganisms to mitigate pollution. This review combines insights from two perspectives: one focusing on the mechanisms, applications, and types of bioremediation, and the other examining the transformative potential of integrating Internet of Things (IoT), Artificial Intelligence (AI), and biosensors ...
Environmental pollution generated the need to search for new environmentally friendly, low-cost, and more efficient environmental clean-up techniques for its removal or reduction. Bioremediation, a branch of environmental biotechnology, is nowadays considered as one of the most promising alternatives.
An increased amount of toxins has collected in the environment (air, water, and soil), and traditional methods for managing these pollutants have failed miserably. Advancement in modern remediation techniques could be one option to improve bioremediation and waste removal from the environment. The increased pollution in the environment prompted the development of genetically modified ...
Environmental pollution and its remediation are one of the major problems around the globe. Broad varieties of pollutants viz. pesticides, hydrocarbons, heavy metals, and dyes, etc. are the key players, which are mainly responsible for environmental pollution. Residual contaminants are also difficult to eliminate. Bioremediation is one of the most efficient technologies for the reduction of ...
Yet, bioremediation with the data-assisted synthetic biology is overlooked. Conversely, bioremediation properties/pathways of a natural enzyme could be optimized by data-assisted assisted enzyme engineering. Air Pollution, CO 2 Fixation, and Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (RUBisCO)
Bioremediation for Environmental Sustainability: Approaches to Tackle Pollution for Cleaner and Greener Society discusses many recently developed and successfully applied bio/phytoremediation ...
These cost-effective and environment-friendly methods of reducing pollutants are called bioremediation. In bioremediation methods, enzymes play the most crucial role. ... contaminated sites: concepts, applications and challenges. Environmental Science and Pollution Research. 2020;27(22):27563-27581. doi: 10.1007/s11356-020-08903-. [Google ...
To accurately gauge environmental pollution degradation rates, it's necessary to consider all aspects of a given operation. Even if degradation microorganisms exist in the environment, contaminants will not be degraded on a regular basis under competitive circumstances. ... Bioremediation research are awaiting the introduction of these gene ...
Also, if the sources of the nutrients are chemicals (synthetic), they can serve as a source of pollution to the environment defeating the initial purpose of bioremediation . 4.5 Bioleaching Bioleaching is the process of utilizing acidophilic microbes to promote the solubilization of heavy metals which are in a solid state from the sediment matrix.
Environmental remediation is crucial for restoring and preserving our planet's ecosystems. Recent centuries of rapid industrialization, urbanization, and agricultural intensification have led to increased air, water, and soil pollution, posing significant threats to biodiversity, public health, and natural systems (Bediako et al., 2023).This introduction explores the latest advancements in ...