
- Common misconceptions
- Set Your Location
- Learn the signs
- Symptoms of autism
- What causes autism?
- Asperger syndrome
- Autism statistics and facts
- Learn about screening
- Screening questionnaire
- First Concern to Action
- Autism diagnostic criteria: DSM-5
- Newly diagnosed
- Associated conditions
- Sensory issues
- Interventions
- Access services
- Caregiver Skills Training (CST)
- Information by topic
- Resource Guide
- Autism Response Team
- Our mission
- Our grantmaking
- Research programs
- Autism by the Numbers
- Fundraising & events
- Field Day For All
- Social fundraising
- Ways to give
- Memorial & tributes
- Workplace giving
- Corporate partnership
- Become a partner
- Ways to engage
- Meet our Partners
- Primeros signos de autismo
- Síntomas del autismo
- ¿Qué causa el autismo?
- Signos de autismo en adultos
- Signos de autismo en mujeres y niñas
- Detección del autismo
- Deteccion temprana
- Criterios de diagnóstico del autismo
- Niveles de autismo
- Síndrome de Asperger
- Select Your Location
Please enter your location to help us display the correct information for your area.

Research roundup: 2024 in review and the road ahead

2024 brought important progress in autism research, with discoveries that deepen our understanding of autism and open new possibilities for improving care. Advances in genetic research, technology and diagnostic tools are paving the way for better outcomes for autistic people and their families.
This momentum will only grow stronger with the passage of the Autism CARES Act of 2024 , which reaffirms a national commitment to addressing the diverse needs of the autistic community. By prioritizing equity, representation and lifelong support, this law will help ensure that more voices are heard and more people receive the care they need.
Dr. Andy Shih, Chief Science Officer, Autism Speaks
Key autism research advancements in 2024
Read what the members of the Autism Speaks Medical and Science Advisory Committee see as the top research trends of 2024:
- New research explored social inequities in autism
Emerging technologies unlocked new possibilities
- Innovations expanded access to autism screening and diagnosis
- Focus shifted to people with profound autism
- Advances in genetic research paved the way for gene-targeted therapies
- Autism subtyping laid the groundwork for personalized care
Autistic-led research gained momentum
What’s next—trends in 2025, new research explored social inequities in autism .

Research this year highlighted the systemic barriers facing the autism community. A study from the Baby Siblings Research Consortium found that infants with older autistic siblings were more likely to be diagnosed with autism if their families were from marginalized communities or had less access to education.
“I believe there’s growing interest in understanding how social determinants of health like poverty or racism affect overall health and well-being,” says Dr. Brian Boyd, William C. Friday distinguished professor in education in the School of Education at University of North Carolina at Chapel Hill. “Understanding how these factors are impacting the ability of autistic people to thrive and have good quality of life is very important.”
Some studies have examined historically underrepresented communities. Research from the University of North Carolina TEACCH Autism Program analyzed 20 years of diagnostic trends among females and found that while diagnosis rates are rising faster than among males, females are still more likely to be diagnosed later in life—often after age 13.
“This kind of research gives us more of a systems-level understanding of what’s impacting autistic people’s outcomes,” explains Dr. Boyd. “By looking at broader structural issues, we can go beyond individual-level interventions and into system-level solutions that have a bigger impact on people’s lives and outcomes.”
Whether it’s things like concentrated poverty or racism, understanding how these factors are impacting the ability of autistic people to thrive and have good quality of life is very important. Dr. Brian Boyd

Technological innovations like generative artificial intelligence (AI), extended reality (XR), virtual assistants and wearable devices are revolutionizing support for people with autism, offering new possibilities in education, employment, social interaction and independent living.
“With the proper development and guidance, these technologies hold the promise of significantly improving accessibility and inclusion across the lifespan,” says Dr. Russell Shilling, former Navy Captain, aerospace experimental psychologist, program officer at the Defense Advanced Research Projects Agency (DARPA) and chief scientific officer of the American Psychological Association (APA).
A study showed that AI chatbots are providing valuable advice about workplace social communication to employed autistic adults, while a meta-analysis found that VR-based interventions are effective in helping children and adolescents with autism improve cognitive, social and emotional skills.
“Generative AI facilitates personalized communication aids, enabling individuals to express themselves more effectively and receive tailored organizational support,” says Dr. Shilling. “When integrated with virtual assistants, these technologies can improve workplace efficiency and support independent living. In educational settings, AI-driven tools can adapt learning materials to individual needs, fostering a more inclusive and effective learning environment.”
Innovations expanded access to autism screening and diagnosis

Advancements in diagnostic tools and methods are paving the way for more accessible autism screening worldwide. Research on gut microbiota revealed distinct microbial profiles in autistic people, showing promise for stool analysis as a quick, accessible autism screening tool.
“If the results of this study can be implemented in real-world contexts, it raises hopes for a cheap, fast and simple screening test for autism that may not require specialists initially,” says Dr. Angelina Kakooza-Mwesige, senior lecturer and pediatric neurologist at the School of Medicine at Makerere University College of Health Sciences in Kampala, Uganda.
Another study found that using the M-CHAT-R/F tool for routine, standardized autism screenings during well-child check-ups is more effective than typical clinical surveillance. Conducting these screenings universally, even in the absence of obvious autism signs, led to increased referrals for diagnosis—highlighting its potential to impact healthcare systems worldwide.
If the results of this study can be implemented in real-world contexts, it raises hopes for a cheap, fast and simple screening test for autism. Dr. Angelina Kakooza-Mwesige
Focus shifted to people with profound autism

New research brought greater visibility to the experiences of people with profound autism, or those with the highest support needs—a group that has historically been excluded from autism research.
“For many years, children with significant cognitive and language delays were treated as a homogeneous group at the far end of the autism spectrum and were often excluded from studies due to the lack of appropriate assessments and effective interventions. However, 2024 saw a shift, with multiple publications specifically addressing this population,” says Dr. Connie Kasari, professor of human development and psychology at the Semel Institute at University of California, Los Angeles.
One study of 344 minimally verbal autistic children revealed a broad range of cognitive and communication abilities, challenging assumptions of homogeneity within this group. Others explored the use of gesture and its role in language outcomes . A thought-provoking essay called for greater integration of people with profound autism into research studies, emphasizing the dire need to reduce marginalization and address their unique needs.
Advances in genetic research paved the way for gene-targeted therapies

Autism genetics research made significant advancements this year, accelerating the development of gene-targeted therapies. In a study anticipated to be published next year, the Autism Sequencing Consortium (ASC) conducted the largest exome study of autism spectrum disorder (ASD) to date, finding 230 autism-related genes—up from 72 genes in the previous 2022 analysis .
“I think we can fairly say that we’re at least at 50% in terms of what we can discover from genome sequencing,” says Dr. Joseph Buxbaum, professor of psychiatry, neuroscience and genomic sciences, co-founder of the ASC, and director of the Seaver Autism Center at Icahn School of Medicine at Mount Sinai. “Compared to a few years ago, we have tripled the number of autism-associated genes. Now, we are beginning to see researchers taking these genetic findings and making very clear advances in terms of therapeutics.”
These discoveries are paving the way for new treatments that are improving quality of life for autistic people. Clinical trials this year have shown that gene-targeted therapies are improving cognitive abilities, communication skills and daily living skills among people with Fragile X syndrome and Angelman syndrome , two rare monogenic forms of autism.
“As we uncover more genetic factors that influence autism presentation and outcomes, autistic people will be better positioned to benefit from personalized healthcare just like everyone else,” says Dr. Shih. “With gene-targeted therapies, we’ll be able to improve the quality of life for people with autism well into adulthood.”
I think we can fairly say that we’re at least at 50% in terms of what we can discover from genome sequencing. Dr. Joseph Buxbaum
Autism subtyping laid the groundwork for personalized care

Emerging research from the SPARK and Simons Simplex Collection research cohorts led to a major genetic discovery, identifying four distinct autism subtypes. Based on genetic and phenotypic data from more than 5,000 autistic children, this study was the first to link autism subtypes to underlying biological pathways, setting the stage for more personalized approaches to care.
While gene-targeted therapies target specific genetic mutations, subtyping takes a broader approach, enabling interventions that are tailored to shared biological and behavioral traits across subgroups.
“Care needs to be personalized to be most effective,” explains Dr. Shih. “Think of it like getting eyeglasses: everyone has a unique prescription to help them see better, and that prescription can change over the course of your life based on your genetics and environment. In the same way, personalized care for autistic people should fit their specific needs and adapt as their needs evolve. Subtyping helps us understand the unique characteristics of autistic people—whether as part of a subgroup or as individuals—so we can deliver the most effective care.”
2024 saw a significant shift toward research led by autistic scientists, bringing unique insights and priorities to the field. For a second year, Autism Speaks offered its predoctoral fellowship for autistic researchers that aims to increase representation in the field and advance diverse areas of study.
“This is going to be the third year that we’re running our program, and we’ve been so impressed by the quality of work, diversity of ideas and areas of investigation among our applicants,” says Dr. Shih.
This growing representation is already driving change. “As we have more research led by autistic researchers, we are shifting some of our research priorities,” says Dr. Boyd. “It’s really helping us understand where there are areas of convergence or agreement among different groups within the autism community. Mental health and aging with autism have emerged as key themes, along with the importance of diversifying the voices that are being represented within autism research.”
One such project , led by Autism Speaks predoctoral researcher Patrick Dwyer , explored perspectives on neurodiversity within the autism and autistic communities. His research highlighted the nuance of these perspectives: even those who embrace the social model of disability agreed on the need for individualized support, particularly around adaptive skills, depression and epilepsy—areas where targeted interventions can make a lasting difference.
As we have more research led by autistic researchers, we are shifting some of our research priorities. Dr. Brian Boyd
As we turn our attention to 2025, the future looks promising:
- The reauthorization of the Autism CARES Act will fundamentally reshape the research agenda, prioritizing work on aging, level 3 autism, communication among those who are minimally verbal and training professionals to deliver quality care.
- The growing voice of autistic researchers will shape priorities to better reflect community needs.
- Advances in gene-targeted therapies and autism subtyping will continue to drive personalized healthcare , offering tailored solutions that meet the unique needs of people with autism.
- Emerging technologies like AI and XR will revolutionize skill building, communication and daily living support for people with autism.
- Research on social determinants of health will drive system-level interventions that address inequities and improve access to support.
- The inclusion of traditionally unrepresented populations , including those who are minimally verbal and from diverse cultural and socioeconomic backgrounds, will ensure that research reflects the full scope of autistic people’s experiences.
With continued collaboration, we can ensure that this research translates into meaningful quality of life improvements for autistic people and their families.
Additional Resources & Tools

Genetic discovery links new gene to autism spectrum disorder
Variants in the ddx53 and other genes on the x chromosome provide genetic clues to male prevalence in asd.
New research published in The American Journal of Human Genetics has identified a previously unknown genetic link to autism spectrum disorder (ASD). The study found that variants in the DDX53 gene contribute to ASD, providing new insights into the genetic underpinnings of the condition.
ASD, which affects more males than females, encompasses a group of neurodevelopmental conditions that result in challenges related to communication, social understanding and behaviour. While DDX53 , located on the X chromosome, is known to play a role in brain development and function, it was not previously definitively associated with autism.
In the study published today, researchers from The Hospital for Sick Children (SickKids) in Canada and the Istituto Giannina Gaslini in Italy clinically tested 10 individuals with ASD from 8 different families and found that variants in the DDX53 gene were maternally inherited and present in these individuals. Notably, the majority were male, highlighting the gene's potential role in the male predominance observed in ASD.
"By pinpointing DDX53 as a key player, particularly in males, we can better understand the biological mechanisms at play and improve diagnostic accuracy for individuals and their families," says senior author Dr. Stephen Scherer, Senior Scientist, Genetics & Genome Biology and Chief of Research at SickKids, and Director of the McLaughlin Centre at the University of Toronto.
"Identifying this new gene as a confirmed contributor to ASD underscores the complexity of autism and the need for comprehensive genetic analysis."
At the same location on the X chromosome, the researchers found evidence that another gene, PTCHD1-AS , might be involved in autism. The study highlights a case where a boy and his mother, both with autism with little support needs, had a specific gene deletion involving the DDX53 gene and parts of PTCHD1-AS .
The study cohort was assembled through an international collaborative effort, involving several renowned clinical and research institutions from Canada, Italy and the U.S. Further analysis of large autism research databases, including Autism Speaks MSSNG and Simons Foundation Autism Research Initiative, identified 26 more individuals with ASD who had similar rare DDX53 variants to the study participants.
"This gene has long eluded us, not previously linked to any neuropsychiatric condition. Our findings support a direct link between DDX53 and autism, which is not only crucial for future clinical genetic testing, but its discovery suggests that the pathway it affects is related to the behavioural traits of autism, opening a whole new area of exploration," says lead author Dr. Marcello Scala, researcher in Medical Genetics at the Istituto Giannina Gaslini, affiliated with the University of Genoa (Department of Neuroscience).
In another paper published today in the same journal, Scherer and lead author Dr. Marla Mendes, a research fellow at SickKids, identified 59 genetic variants on the X chromosome significantly associated with ASD. The variants were found in genes linked to autism, including PTCHD1-AS (near to DDX53 ), DMD , HDAC8 , PCDH11X, and PCDH19 beside novel ASD-linked candidates ASB11 and ASB9 . Additionally, the FGF13 gene was highlighted as being related to ASD, with sex-specific differences, adding more evidence to the role of sex chromosomes in the condition.
"These findings provide new insights into the biology of the X chromosome in ASD, providing additional evidence for the involvement of certain genes like DDX53 and FGF13 , and suggesting they should be investigated further," says Scherer.
The team notes that the absence of a similar gene like DDX53 in commonly used mouse models may require future researchers to reconsider how they study ASD. Since it lacks a functional equivalent in these models, findings in DDX53 cannot be easily replicated.
"Insights from this study could significantly influence the design and interpretation of autism research, particularly in developing new models. Identifying these variants is an important step towards developing more precise diagnostics and therapeutics for patients and families with ASD," says Scherer.
Scherer also added "both studies provide even more evidence that complex neurobehavioral conditions like autism can sometimes have simple biologic (genetic) underpinnings."
The study was funded by the University of Toronto McLaughlin Centre, Autism Speaks, Autism Speaks Canada, Ontario Brain Institute, the Italian Ministry for Education, University and Research and SickKids Foundation. Additional funding was provided by National Institutes of Health and the California Center for Rare Diseases at UCLA.
- Birth Defects
- Gene Therapy
- Personalized Medicine
- Learning Disorders
- Disorders and Syndromes
- Child Development
- Autistic spectrum
- Bipolar disorder
- Psychiatric service dog
- Adult attention-deficit disorder
- Asperger syndrome
- Molecular biology
Story Source:
Materials provided by The Hospital for Sick Children . Note: Content may be edited for style and length.
Journal Reference :
- Marcello Scala, Clarrisa A. Bradley, Jennifer L. Howe, Brett Trost, Nelson Bautista Salazar, Carole Shum, Marla Mendes, Miriam S. Reuter, Evdokia Anagnostou, Jeffrey R. MacDonald, Sangyoon Y. Ko, Paul W. Frankland, Jessica Charlebois, Mayada Elsabbagh, Leslie Granger, George Anadiotis, Verdiana Pullano, Alfredo Brusco, Roberto Keller, Sarah Parisotto, Helio F. Pedro, Laina Lusk, Pamela Pojomovsky McDonnell, Ingo Helbig, Sureni V. Mullegama, Emilie D. Douine, Rosario Ivetth Corona, Bianca E. Russell, Stanley F. Nelson, Claudio Graziano, Maria Schwab, Laurie Simone, Federico Zara, Stephen W. Scherer. Genetic variants in DDX53 contribute to autism spectrum disorder associated with the Xp22.11 locus . The American Journal of Human Genetics , 2024; DOI: 10.1016/j.ajhg.2024.11.003
Cite This Page :
Explore More
- Can the Heart Heal Itself? New Study Says It Can
- Tinkering With 'Clockwork' Mechanisms of Life
- Quantum Teleportation Over Busy Internet Cables
- Mysteries of Icy Ocean Worlds
- Safer Spuds: Removing Toxins from Potatoes
- Gruel Eaten by Early Neolithic Farmers
- Dark Energy 'Doesn't Exist'
- Nerve Regeneration After Spinal Cord Injury
- Laser-Based Artificial Neuron: Lightning Speed
- Large Hadron Collider Regularly Makes Magic
Trending Topics
Strange & offbeat.
- Open access
- Published: 18 December 2024
Exploring autism spectrum disorder and co-occurring trait associations to elucidate multivariate genetic mechanisms and insights
- Karoliina Salenius 1 na1 ,
- Niina Väljä 1 na1 ,
- Sini Thusberg 1 ,
- Francois Iris 3 ,
- Christine Ladd-Acosta 4 ,
- Christophe Roos 5 ,
- Matti Nykter 1 , 6 ,
- Alessio Fasano 7 , 8 ,
- Reija Autio 9 na1 ,
- Jake Lin 1 , 10 na1 on behalf of
the GEMMA study
BMC Psychiatry volume 24 , Article number: 934 ( 2024 ) Cite this article
170 Accesses
Metrics details
Autism spectrum disorder (ASD) is a partially heritable neurodevelopmental trait, and people with ASD may also have other co-occurring trait such as ADHD, anxiety disorders, depression, mental health issues, learning difficulty, physical health traits and communication challenges. The concomitant development of ASD and other neurological traits is assumed to result from a complex interplay between genetics and the environment. However, only a limited number of studies have performed multivariate genome-wide association studies (GWAS) for ASD.
We conducted to-date the largest multivariate GWAS on ASD and 8 ASD co-occurring traits (ADHD, ADHD childhood, anxiety stress (ASDR), bipolar (BIP), disruptive behaviour (DBD), educational attainment (EA), major depression, and schizophrenia (SCZ)) using summary statistics from leading studies. Multivariate associations and central traits were further identified. Subsequently, colocalization and Mendelian randomization (MR) analysis were performed on the associations identified with the central traits containing ASD. To further validate our findings, pathway and quantified trait loci (QTL) resources as well as independent datasets consisting of 112 (45 probands) whole genome sequence data from the GEMMA project were utilized.
Multivariate GWAS resulted in 637 significant associations ( p < 5e-8), among which 322 are reported for the first time for any trait. 37 SNPs were identified to contain ASD and one or more traits in their central trait set, including variants mapped to known SFARI ASD genes MAPT , CADPS and NEGR1 as well as novel ASD genes KANSL1 , NSF and NTM , associated with immune response, synaptic transmission, and neurite growth respectively. Mendelian randomization analyses found that genetic liability for ADHD childhood, ASRD and DBT has causal effects on the risk of ASD while genetic liability for ASD has causal effects on the risk of ADHD, ADHD childhood, BIP, WA, MDD and SCZ. Frequency differences of SNPs found in NTM and CADPS genes, respectively associated with neurite growth and neural/endocrine calcium regulation, were found between GEMMA ASD probands and controls. Pathway, QTL and cell type enrichment implicated microbiome, enteric inflammation, and central nervous system enrichments.
Conclusions
Our study, combining multivariate GWAS with systematic decomposition, identified novel genetic associations related to ASD and ASD co-occurring driver traits. Statistical tests were applied to discern evidence for shared and interpretable liability between ASD and co-occurring traits. These findings expand upon the current understanding of the complex genetics regulating ASD and reveal insights of neuronal brain disruptions potentially driving development and manifestation.
Multivariate GWAS resulted in 637 significant ASD associations ( p < 5e-8), among which 322 are reported for the first time.
The novel associations mapped to known SFARI ASD genes CADPS , MAPT and NEGR1 and novel ASD genes KANSL1 , NSF and NTM , associated with immune response, synaptic transmission, and neurite growth, potentially driving the gut brain-barrier hypothesis underpinning ASD development.
CuONPs induce co-occurrence of autophagy activation and autophagic flux blockade.
Mendelian randomization analyses found that genetic liability for ASRD and DBT have causal effects on the risk of ASD while genetic liability for ASD have causal effects on the risk of ADHD, BIP, WA, MDD and SCZ. Bidirectional genetic liability causal effects were confirmed between ASD and ADHD childhood.
Peer Review reports
Introduction
ASD spectrum disorders (ASD) is an umbrella term for a group of heterogeneous neurodevelopmental traits that manifest in early childhood. ASD is a complex disorder with both genetic and environmental risk factors [ 10 , 30 , 45 ]. The diagnosis of ASD is based on its key characteristics including difficulties in social communication and interaction, restricted and repetitive behaviors, hyperactivity and divergent responses to sensory inputs. The most common co-occurring traits in autistic persons are attention deficit hyperactivity disorder (ADHD), ADHD childhood, anxiety, bipolar (BP), depression, epilepsy, obsessive compulsive disorders (OCD) and stress related traits, all of which share overlapping diagnostic attributes and challenging symptoms with ASD [ 30 , 57 ]. According to US data, autistic children tend to fare less well in educational attainment (EA) and about one in three have a reduced intellectual ability, as defined by intelligence quotient (IQ less than 70) [ 4 , 68 ]. Some children with ASD having higher IQ scores also comparatively experience harder academic struggles due to co-occurring traits and difficulties in social interactions [ 3 ].
Together with recent advances in genomics technology and pivotal support from the engaged ASD community, 1,162 genes are currently implicated with ASD development and these are curated in the SFARI [ 2 , 19 , 52 ] gene module. These genes, with varying degrees of effect, are scored using the Evaluation of ASD Gene Link Evidence (EAGLE) framework [ 61 ]. Surprisingly, while it is known that common variants contribute to most of the genetic background [ 18 ], only a few robust genetic associations have been recently reported. Most of these are attributed to the landmark study conducted by Grove and colleagues, employing a large Danish cohort with 18,381 ASD cases and 27,969 controls, where 12 significant variant associations were reported [ 19 ].
Given that there is overlap in symptoms between ASD and ADHD, recent genetics studies found shared genetic factors underlying ASD and ADHD [ 40 , 41 , 50 ], with partial concordance between bidirectional colocalization single nucleotide variants (SNPs). However, these studies were limited to general ADHD (onset age 10+), and not childhood ADHD. Astoundingly many (47% median) autistic children have reported one or more gastrointestinal (GI) symptoms [ 5 ]. Recently, there have been promising results that link microbiome disruption and diversity [ 44 ] as a novel contributing factor to ASD. While Grove and colleagues found that 7 of the 12 ASD SNP associations have similar significance towards EA and psychosis traits depression and schizophrenia [ 19 ], still little is known concerning the joint liability and the shared genetic mechanisms between ASD and ASD co-occurring traits including ADHD, ADHD childhood, anxiety-stress related disorder (ASRD), bipolar, disruptive behavior disorder (DBD), EA, epilepsy, inflammatory bowel disease (IBD), major depression, obsessive compulsive disorder (OCD) and schizophrenia (SCZ). Respectively, the 11 co-occurring trait summary statistics are retrieved from large reputable cohorts, listed in Table 1 and Supplementary Table 1.
To attenuate the genetic knowledge gaps in ASD and expand the exploration of potential shared co-occurring trait genetic associations, this study performed multivariate genome-wide association study (GWAS) with summary statistics from ASD and 11 co-occurring traits from large reputable cohorts. To achieve this, colocalization (coloc) was systematically applied to test the robustness between the shared variants and traits [ 75 ]. Mendelian randomisation (MR) was further applied, using the multivariate variants and the essential traits, to assess liability relationships between ASD and the selected co-occurring traits [ 6 , 55 ]. This study seeks to further clarify functional, regulatory and tissue type differentiation with enrichment and integration of quantified trait loci (QTL) while validating our key findings with independently sequenced genomes from the GEMMA cohort [ 70 ].
Methods and materials
GWAS summary statistics for ASD and ADHD were collected from the Psychiatric Genomics Consortium (PGC) and iPSYCH [ 49 , 65 ] studies. Education attainment [ 47 ] summary file was collected from the Social Science Genetic Association Consortium (SSGAC). Additional ASD co-occurring traits, selected based on LDSC (LD Score Regression) genetic correlation ( p -value < 0.05) with ASD, include ADHD childhood, bipolar (BP), anxiety-stress disorder (ASRD), disruptive behaviour (DBD), major depression (MDD) and schizophrenia (SCZ), with sample sizes ranging from 31,890 − 765,283 are shown in Table 1 (additional details including doi references listed Supplementary Table 1). To estimate potential sample overlaps, pairwise LDSC intercepts with ASD are calculated and reported in Supplementary Table 1. Summary statistics are joined, yielding 4,525,476 SNPs, and applied in a multivariate GWAS setting. Follow-up analysis includes decomposition aiming to detect the most important traits while colocalization and Mendelian randomisation analysis are conducted to explore shared liability as shown in Fig. 1 .
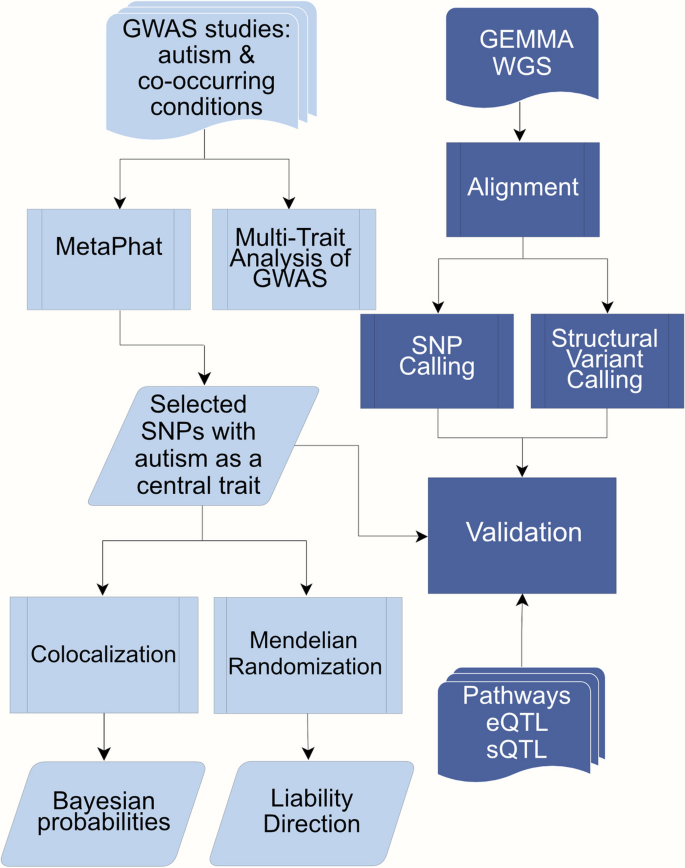
Workflow for the analyses conducted in the study. Multivariate GWAS was performed on selected GWAS studies including ASD and 8 co-occurring traits: ADHD, ADHD childhood, bipolar, anxiety, disruptive behaviour, educational attainment, major depression and schizophrenia. 37 SNPs were selected and evaluated with Colocalization and Mendelian Randomization. Further validation of these SNPs utilized pathway and EBI eQTL/sQTL catalogs as well as the GEMMA -study. The GEMMA whole genome sequencing (WGS) processing included variant calling to infer structural and single nucleotide variants (SVs and SNVs) present in the samples
Multivariate GWAS and determination of central traits
Multivariate GWAS on ASD and ASD co-occurring traits were performed using metaPhat/metaCCA software that performs multivariate analysis by implementing Canonical Correlation Analysis (CCA) for a set of univariate GWAS summary statistics [ 12 , 36 , 58 ]. The objective of metaCCA is to find the optimal genetic effect combination that is maximally correlated with a linear combination of the trait variables. ASD multivariate central traits are identified by MetaPhat decomposition based on iterative tracing of p -values (p) from trait subsets (relative to 5e-8) and Bayesian Information Criterion (BIC) [ 62 ] representing model fit. Essentially, driver trait(s) are the subsets of the multivariate association that drives the p-value, and without the drivers, the multivariate association is no longer significant ( p > 5e-8). Similarly, as the decomposition processing is exhaustive (iterates from k to 1), an optimal subset is identified by comparing BIC values [ 36 ]. For simplicity, the central traits are the union of the driver and optimal BIC traits. Multi-Trait Analysis of GWAS (MTAG) [ 71 ], a high performance multivariate-GWAS that addresses sample overlap, is additionally performed for validation.
Genetic annotations, pathway enrichment and validation
SFARI Base Gene resource, GeneCards and GWAS catalog were used to assess the novelty of variants and genes associated with ASD [ 2 , 39 , 60 ]. snpXplorer was applied towards SNP annotation [ 69 ]. Reactome and WikiPathway databases pathway enrichments were evaluated with the Enrichr tool [ 31 ]. Human organ and cell type systems enrichment analysis, encompassing 1,466 tissue-cell type and single-cell RNAseq panels, was conducted using WebCSEA [ 13 , 33 ]. eQTL and sQTL were assessed within the QTL catalog, via FIVEx portal [ 32 ].
Colocalization analyses
Colocalization was performed for the selected multivariate ASD SNPs to assess if the associated variants in the locus are shared genetically between ASD and the 8 co-occurring related ASD traits to account for erroneous results that may follow from analyzing individual SNPs. Errors can occur when a SNP associated with trait 1 and trait 2 are in linkage disequilibrium (LD). The analyses were performed using the R package coloc [ 20 , 33 ].
The colocalization analysis was conducted using the absolute base factor colocalization method (coloc.abf), which is a Bayesian colocalization analysis method. A region size window of 100KB (± 50 KB flanking the SNP position) was selected to comprehensively span potential LD and regulatory elements [ 53 ]. The different hypotheses tested include: H0 (no liable variant), H1 (liable variant only for trait 1), H2 (liable variant only for trait 2), H3 (two separate liable variants), H4 (common liable variant shared between the traits). As recommended [ 74 ], default setting prior probability thresholds were applied: 1e-4 for H1, H2 and H3 and 1e-5 for H4 while posterior probability (H4 > 90%) is conservatively applied to estimate shared liability.
Mendelian randomization analyses
Mendelian Randomization analyses (MR) was conducted on the selected multivariate GWAS SNPs based on their assigned central traits, to explore the liability, direction and independent (reverse causation) relationships between ASD and its related traits [ 51 ]. Instrumental strengths, approximated with F1 score > 10, were calculated using SNP effect and standard error values [6, 49]. To account for the potential biases due to participant overlap between cohorts, the lower bound (95% confidence interval) of the F1 was calculated [ 9 ]. The analyses were performed using the platform TwoSampleMR [ 6 ].
Whole genome sequencing
The results were validated using yet unpublished data from the EU Horizon2020 GEMMA research project with genotype variant calls in 112 (49% female) WGS samples with 45 ASD probands (42% female) from the GEMMA prospective cohort [ 70 ]. These samples, assayed on whole blood and collected during enrollment, were sequenced with 30-40X coverage on Illumina NovaSeq 6000 platform. Data was aligned to GRCh38 reference genome using bwa mem v0.7.17 [ 34 ] and reads were sorted and duplicates marked with samtools v1.12 [ 35 ]. Quality control was performed with omnomicsQ -software [ 20 ]. For variant calling DeepVariant v1.4.0 [ 54 ] was utilized and variants were annotated with Variant Effect Predictor [ 43 ] version 112.0.
Statistical analysis
All statistical analyses were performed using R 4.2.2 software and available as R markdown results in the github project ( https://github.com/jakelin212/mvasd_gwas ). Genome-wide association is called on the standard and strict p-value threshold of 5e-8 (-log10 7.3), to account for multiple testing based on the assumption of about 1-million independent tests [ 56 ]. To assess SNP allele proportional differences for validation, the phi coefficient is computed, and statistical significance was determined using Chi-square test. Fisher’s exact test was used when Chi-square assumptions were not met. Bonferroni correction is assessed to account for multiple testing of the multivariate GWAS involving 9 traits ( p < 5.5e-9; -log10(p) > 8.25).
GWAS summary statistics
GWAS summary statistics for ASD and ADHD were collected from the PGC and iPSYCH [ 49 , 65 ] studies. Education attainment [ 47 ] summary file was collected from the Social Science Genetic Association Consortium (SSGAC). Altogether, using summary statistics, 11 ASD co-occurring traits were assessed for genetic correlation with the landmark ASD study [ 19 ], the largest genetic correlation values, as computed by LDSC [ 8 ], were between ASD and ADHD (rg = 0.535), followed by MDD (rg = 0.505) and ADHD childhood (rg = 0.478). Shown in Table 1 below, 8 traits are shown to be genetically correlated with ASD ( p < 0.05) and additional details of all traits are shown in Supplementary Table 1.
Multivariate ASD central trait SNPs, pathway and organ tissue enrichment
Multivariate GWAS was performed with ASD together with its genetically correlated traits, ADHD, ADHD childhood, ASRD, bipolar, DBD, EA, MDD, and SCZ (Table 1 ) and 637 ( p < 5e-08) SNP associations were found, including 322 variants that are reported for the first time for any trait (Supplementary Table 6) according to GWAS catalog. Two associations (rs2388334 and rs1452075) intersected with the twelve associations identified in the landmark common genetic variants of ASD study [ 19 ]. When assessed at the gene level, all 12 were concordant (as indicated in STable 6). Decomposition implemented in MetaPhat, using stepwise tracing of p -value and Bayesian information criteria (BIC) contributions [ 36 , 62 ], identified 37 ASD central trait SNPs where 16 were identified with multivariate GWAS approach (all SNPs p < 5.5e-09; min (-log10(p) 8.67), listed in Supplementary Table 2). These 37 multivariate ASD SNPs, 17 of which had previously been reported in existing GWAS studies, mapped to 35 genes (Table 2 ) and confirmed that 8/35 ( ARHGAP32 , CADPS , CUL3 , KANSL1 , MACROD2 , MAPT , MSRA and NEGR1 ) are known curated SFARI genes, with ASD susceptibility EAGLE scores < = 3 (indicating limited evidence) [ 61 ]. The variant rs538628 within the NSF gene, a regulator of AMPA receptor endocytosis and critical for mediating glutamatergic synaptic transmission [ 25 ], along with the variant rs62061734 in the MAPT gene, are identified to associate with the optimal central traits of ASD, EA and SCZ ( MAPT variant rs62061734 p = 3.98e-31, NSF variant rs538628 p = 1.99e-27, Supplementary Table 2, trace plots are provided in supplementary data). Notably, NSF was previously implicated only in mouse models exhibiting ASD-like behaviors [ 76 ]. Shown in the same table, MTAG [ 71 ] multivariate GWAS validation was performed to address iPSYCH cohort sample overlaps between ADHD and ASD [ 40 , 41 ] subjects where similar results were found ( MAPT variant rs62061734 p = 1.99e-20, NSF variant rs538628 p = 5.37e-18).
Shown in Supplementary Table 7, Fig. 2 e and Supplementary Fig. 3, pathway enrichment using the 35 associated genes was performed with Enrichr [ 31 ]. Nervous systems development (GO:0007399) was found to be the most significant ( p = 1.73e-08) while neural and microtubule structural related pathway hits from Reactome [ 16 ] and WikiPathways [ 46 ] featured pathways were Inclusion Body Myositis ( MAPT and PSEN1 , p = 1.27e-04) and COPII-mediated Vesicle Transport ( NSF and SERPINA1 , p = 4.69e-03). Enrichment analysis was conducted using the WebCSEA tool, which identified statistically significant associations (Fig. 2 f, p < 1e-03) with the following human organ systems: digestive, nervous, sensory, lymphatic, and respiratory. As shown in Supplementary Fig. 4, the most enriched tissue types are related to cerebrum, cortex, intestine and blood related components discerned from 1,355 tissue-type (TS) as well as data from the human brain single cell project [ 33 ].
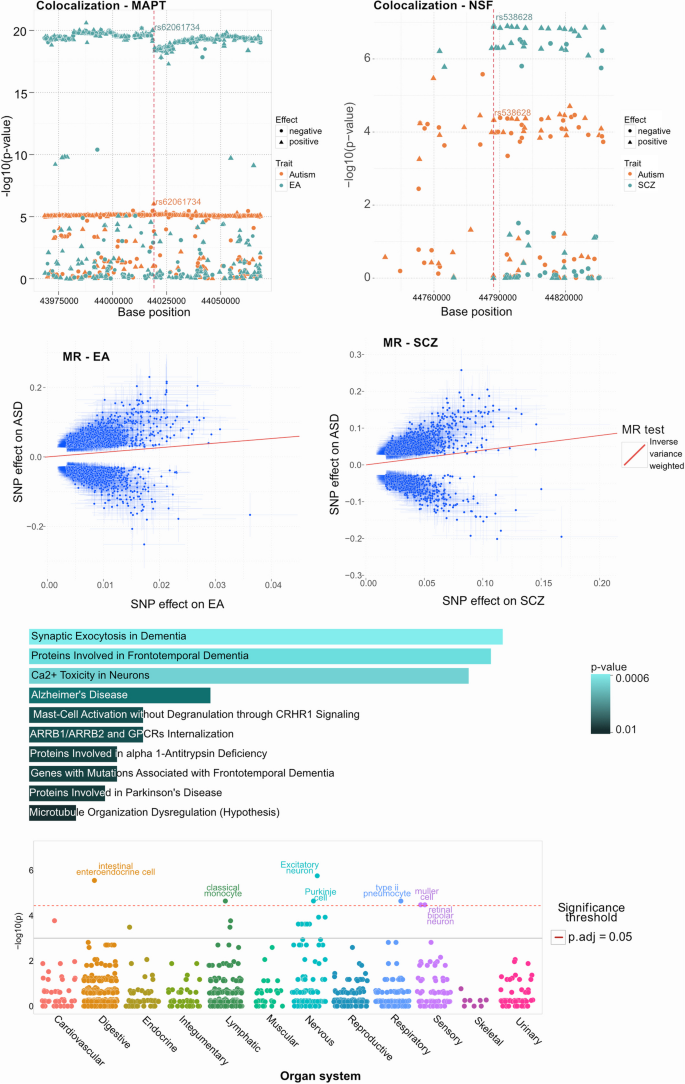
Results from the post GWAS analysis of the 37 selected SNPs. a,b ) Colocalization processing using the original summary statistics of ASD and EA for (a) rs62061734 ( MAPT , failed colocalization with H4 probability 8.19%, p = 0.09), ASD and NSF for (b) rs538628 ( NSF , SCZ passed colocalization with H4 probability 94%, p = 1.1e-05), depicting supporting regional SNPs (x-axis) and their negative log10 p -value (y-axis) and effect direction (circles negative, triangles positive). c,d ) Mendelian randomization (MR) results using inverse variance weighted (IVW) -method for association of ASD SNP effects (y-axis) and c) EA and d) SCZ effects (x-axis). e ) Pathway analysis for the genes associated with the selected SNPs shows enrichment in processes related to neurons using Reactome database. The length of the bar represents the significance of that specific gene-set or pathway and the color indicates the significance of the pathway. Details of the pathways and genes with their associated p-values are listed in Supplementary Table 8. f ) Organ system enrichment was applied using WebCSEA, using the selected 37 multivariate gene associations and found enrichment ( p < 1e-03) with the ASD relevant digestive, nervous and sensory organ systems as well as lymphatic and respiratory systems
Colocalization analysis was conducted on the 37 multivariate SNP associations identified to contain ASD as a central trait. The comparative analysis was performed on the relevant mapped gene window, from start to end while adding 25 KBs on both ends to cover regulating and promoter regional elements. For the two SNPs that did not map to a gene, the window size used for the colocalization analysis was 100 KB (± 50 KB), estimated and derived from the gene median length of 24KB [ 17 ]. Additional information concerning the number of regional LD adjusted SNPs applied to the colocalization test is shown in Supplementary Table 3.
A total of 19/37 SNPs showed strong evidence for a common liability variant with ASD (H4 > 90%, details shown in Supplementary Table 3) and the traits having common ASD liable variants included EA (9), SCZ (6), BP (2), ADHD (1) and ASRD (1). Notably, SNP rs62061734, mapping to the MAPT gene and rs538628, mapping to the NSF gene had H4 of 99% for EA and SCZ, respectively (shown in Fig. 2 a-b) while SNP rs568828, mapping to the NTM gene had H4 of 99% for SCZ and ADHD (Supplementary Table 2).
Mendelian randomization analysis was conducted for the 8 traits genetically correlated (Table 1 ) with ASD. The lead SNPs, with F1 scores > 25 (listed in Supplementary Table 4, where > 10 is considered strong [ 48 ] were found to lend significantly increase probability of ASD ( p < 0.001 both Inverse Variance Weighted (IVW)-method and MR-Egger (EA and SCZ are shown in Fig. 2 c-d), accounting for horizontal pleiotropy and multiple testing with Bonferroni correction of 8 traits). Based on TwoSampleMR Steiger [ 22 ] test for directionality and shown in supplementary Table 4 A, genetic liability to ADHD childhood ( p < 2.44e-116), ASDR ( p < 9.08e-166) and DBD ( p < 1.20e-45) were found to have causal effects on the risk of ASD. Shown in Supplementary Table 4B, genetic liability to ASD ( p < 4.1e-115) were found to have causal effects on the risk of ADHD, ADHD childhood, BIP, EA, MDD and SCZ. The related MR results adhere to the MR-STROBE guidelines [ 64 ].
To assess the impact of the reported multivariate associations on expression (eQTL) and splicing regulatory quantitative trait loci across tissues, the majority (22/37 eQTL, 24/37 sQTL, details listed in Supplementary Table 9) of the associations found are cited in the EBI QTL Catalog [ 28 ] where they associate (adjusted p < 0.05) with adipose, brain and neuron tissues. Furthermore, filtering on GeneCards [ 60 ] curations, the presented ASD central genes are enriched with systems related to gut, microbiome, intestinal immune, enteric nervous and central nervous systems (Supplementary Table 5).
Additionally, the distribution of these ASD-central trait related SNPs in 112 (49% females; 45 ASD probands (42% females) prospective from-birth GEMMA [ 70 ] cohort participants was investigated. SNP distribution differences were for variant rs568828, mapped to NTM and rs62243489, mapped to CADPS . The NTM gene, previously associated with emotional learning deficits in murine models [ 42 ], encodes neurotrimin, while CADPS encodes a neural/endocrine-specific membrane protein regulating calcium. The NTM SNP (rs568828) was present in 42 of 45 probands (92%) compared to 100% of controls (67 of 67). In contrast, CADPS SNP (rs62243489) was found in 19/67 controls (28%) and enriched in 21/45 probands (47%). As listed in Supplementary Table 8, the phi coefficient for NTM between probands and controls was 0.2 ( p = 0.062), while for CADPS , it was − 0.19 ( p = 0.047). When stratified by sex, the phi coefficient for NTM in males was 0.15 ( p = 0.456) and 0.27 ( p = 0.040) in females. For CADPS , the phi coefficients were − 0.18 ( p = 0.182) in males and − 0.20 ( p = 0.140) in females. Notably, the NEGR1 gene (variant rs6699841), involved in neuron growth regulation, showed a phi coefficient of −0.27 ( p = 0.040) in males (24/26 cases; 22/31 controls), while in females, the coefficient was 0.26 ( p = 0.084; 12/19 cases; 31/36 controls). For the variant of NEGR1 , the opposing phi directions between sexes resulted in a phi coefficient of −0.01 ( p = 0.908) in the full dataset. In addition, logistic regression was performed for the specific variants of NTM , CADPS and NEGR1 . The results were not significant for the full cohort (adjusted for sex) or in models stratified by sex.
Using multivariate statistical learning approaches, this study constitutes the largest and most comprehensive genetically correlated multi-trait GWAS analysis with summary statistics performed on ASD and its genetically correlated traits; ADHD, ADHD childhood, ASRD, bipolar, DBD, EA, MDD, and SCZ to explore the underpinnings driving the complexities in ASD. 37 associations containing ASD as a central trait were discovered, with 16 of these associations were detected only due to the increased statistical power of this multivariate GWAS analysis (lowest univariate summary statistics p-value from all traits > 5e-08, and 12/16 confirmed with the MTAG tool [ 71 ], Supplementary Table 2). Interestingly, a previous study using electronic health records of covering nearly 5,000 ASD cases found three subclusters of comorbidity trajectories, first characterized by seizures, then auditory disorders/infections and the third cluster by psychiatric disorders. Due to the complexity of ASD development, a fourth group was described as could not be further resolved. The presented subclusters potentially align well with our ASD central trait sets pertaining to SCZ signals with seizures and psychiatric disorders such as ADHD and intellectual development underpinning EA [ 15 ]. Enrichment analysis confirmed that the multivariate ASD association results are related to neuron and gut tissues and developmental pathways as well as inflammation and microbiome domains, further underscoring the intersection of genome and microbiome as well as supportive of the gut-brain axis hypothesis associated to ASD [ 11 , 44 ]. Surprisingly, genetic correlation performed on LDSC indicated that ASD and IBD are not related (Supplementary Table S1 , rg = −0.059; p = 0.44), a recent report highlighted potential evidence for comorbidity between parental, particularly maternal preexisting IBD onsets and their children developing ASD [ 59 ]. Using the multivariate ASD central trait gene sets, based on comprehensive human tissue cell type and single cell data [ 13 , 33 ] analysis, enrichments were detected with digestive, nervous, and sensory organ systems (Fig. 2 f). At the tissue cell type level and further supporting the gut-brain axis and blood brain barrier, the analysis detected enriched ASD relevant signals related to brain, adipose and gut eQTL/sQTL (Supplementary Table 9) tissue panels.
Overall, the identified ASD traits passed MR with strong F1 measures and significantly contributed to improve the future construction of meta psychiatric based ASD polygenic scores [ 27 ], shown to improve prediction relative to standard PRS in other complex traits such as coronary heart disease and type 2 diabetes [ 37 , 67 ]. The MR results were consistent after calculation of lower bound F1 (all scores > 25, Supplementary Table 4), computed to consider potential biases from cohort sample overlaps [ 9 ]. MR Steiger tests for directionality revealed that genetic liability to ASDR and DBD were found to have causal effects on the risk of ASD. These multivariate ASD associations mapped to genes, including MAPT and NSF which are known to involved in biological pathways linked to neural disorders such as infantile epilepsy [ 66 ] and Parkinson’s Disease [ 7 , 14 ]. Interestingly, colocalization tests for the MAPT region indicated shared genetic risk between only EA and ASD (H4 99%), while that for the NSF gene did not associate with EA, instead associated with SCZ (H4 94%), suggesting intra region heterogeneity that demands future investigation. With respect to ASD, the KANSL1 , BNIP3 , CADPS and NEGR1 genes have been implicated with immune and microbiome features [ 11 ] and behavioral developments [ 63 ]. Similarly, a recent study from Arenella and colleagues reported genetic factors between ASD and various immune phenotypes including KANSL1 associating with lymphocyte counts as well as MAPT associating with eosinophil counts, further supporting the role of the inflammation pathway in ASD development [ 1 ].
The most common traits in our set of 37 associations that passed colocalization with ASD were EA (9), SCZ (6) and BP (2). It is known that the diagnosis for ASD and ADHD, particularly ADHD manifestation in young children, is similar with symptomatic issues concerning hyperactivity and attention span [ 29 ]. While a previous study has performed comparison of genetic and functional enrichment of associations between ASD and ADHD [ 50 ] GWAS resources, this study further complements their results by inclusion of other ASD co-occurring traits, including ADHD and ADHD childhood as well as EA. Interestingly, ASD and ADHD have both been linked with dysbiosis disruption in microbiome composition and function, gastrointestinal and bowel habits issues [ 44 ].
As part of validation, clustering and distribution proportion differences based on the ASD identified SNP associations were detected between probands and non-autistic subjects on genomes from the GEMMA cohort [ 70 ]. Our validation results were performed on the (112, 45 ASD probands) samples currently available in GEMMA. Notably, NEGR1 (rs1432639), a neuronal growth regulator known to associate with migraine, depression and seizures [ 24 , 26 , 73 ], the significant phi coefficients were negative for males and positive for females. Interestingly, a previous study pertaining to prenatal stress found increased NEGR1 expression in the hippocampus of female rats but not in males [ 72 ]. To improve on the specificity and clinical value of the identified traits, a follow-up application of MR with specific expression/protein quantitative loci (tissue/cell type e/pQTL as applied in T1D drug candidate discovery [ 21 ] with genes such as CADPS , NTM and NEGR1 could further reveal molecular and translational insights towards ASD heterogeneity including the high vulnerability subgroup characterized by seizures [ 15 ]. While the validation statistical power was limited by the relatively small sample size, nevertheless the independent and deep sequencing data has allowed the harvesting of interesting observations concerning the distribution of ASD-central trait associations in probands as compared to controls. In addition, the GEMMA validation results should be taken with caution as the population structures (PCs) were not included as covariates due to availability. The upcoming release of additional omics data from GEMMA and other studies, including longitudinal microbiome, metabolome, and methylation datasets, will significantly increase statistical power and enable more detailed temporal analyses. The data will help confirm molecular changes along the gut-brain axis, shedding light on the genetic patterns that contribute to the heterogeneity, development, and comorbidities of ASD. Another limitation of our multi-trait GWAS is that the selection of ASD co-occurring traits is not exhaustive; given the complexity of ASD development, there may be other genetically correlated traits that have not yet been tested at the appropriate population level, warranting consideration and inclusion in future studies. The MR Steiger results on causality need to be taken with caution as unmeasured confounding effects may distort the exposure genetic liability relative to the outcome [ 38 ].
Our study represents the largest multivariate GWAS on ASD to date, combining ASD with eight genetically correlated trait GWAS summaries. We performed systematic decomposition to identify novel genetic associations related to ASD and ASD co-occurring traits. Mendelian randomization testing revealed that genetic liability for ADHD childhood, ASRD and DBD has causal effects on the risk of ASD. Colocalization analysis further confirmed shared genetic risks with ASD, showing enrichment patterns in brain tissues and cell types associated with neurodevelopment, and lending additional support to the gut-brain axis hypothesis.
Data availability
All data generated or analyzed during the study are included and additionally available upon request. For scripts, please see: https://github.com/jakelin212/mvasd_gwas
Arenella M, Fanelli G, Kiemeney LA, Grainne McAlonan DG, Murphy, and Janita Bralten., Brain., behavior, & immunity - health vol. 34 100698. 3 Nov, https://doi.org/10.1016/j.bbih.2023.100698
Arpi MN, Torres, Ian Simpson T. SFARI genes and where to find them; modelling Autism Spectrum Disorder Specific Gene expression dysregulation with RNA-Seq Data. Sci Rep. 2022;12(1):10158.
Article CAS PubMed PubMed Central Google Scholar
Ashburner J, Ziviani J, and Sylvia Rodger. Surviving in the mainstream: capacity of children with Autism Spectrum disorders to perform academically and regulate their emotions and behavior at School. Res Autism Spectr Disorders. 2010;4(1):18–27.
Article Google Scholar
Baio J, Wiggins L, Christensen DL, Maenner MJ, Daniels J, Warren Z, Kurzius-Spencer M, et al. Prevalence of Autism Spectrum Disorder among children aged 8 years - Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2014. Morbidity Mortal Wkly Rep Surveillance Summaries. 2018;67(6):1–23.
Google Scholar
Boorstein HC. Gastrointestinal and feeding problems in Young children: a comparison between Autism Spectrum disorders and other Developmental delays. University of Connecticut; 2008.
Bowden J, Smith GD, and Stephen Burgess. Mendelian randomization with Invalid instruments: Effect Estimation and Bias Detection through Egger Regression. Int J Epidemiol. 2015;44(2):512–25.
Article PubMed PubMed Central Google Scholar
Brion JP, Octave JN, Couck AM. Distribution of the Phosphorylated Microtubule-Associated protein tau in developing cortical neurons. Neuroscience. 1994;63(3):895–909.
Article CAS PubMed Google Scholar
Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL, Neale BM. LD score regression distinguishes confounding from polygenicity in genome-wide Association studies. Nat Genet. 2015;47(3):291–95.
Burgess S, Davies NM, and Simon G. Thompson. Bias due to participant overlap in two-sample mendelian randomization: Burgess Et Al. Genet Epidemiol. 2016;40(7):597–608.
Chaste P, and Marion Leboyer. Autism risk factors: genes, Environment, and gene-environment interactions. Dialog Clin Neurosci. 2012;14(3):281–92.
Cheng L, Wu H, Chen Z, Hao H, and Xiao Zheng. Gut microbiome at the crossroad of genetic variants and Behavior disorders. Gut Microbes. 2023;15(1):2201156.
Cichonska A, Rousu J, Marttinen P, Kangas AJ, Soininen P, Lehtimäki T, Raitakari OT, et al. metaCCA: Summary statistics-based multivariate Meta-analysis of Genome-Wide Association Studies Using Canonical Correlation Analysis. Bioinformatics. 2016;32(13):1981–89.
Dai Y, Hu R, Liu A, et al. WebCSEA: web-based cell-type-specific enrichment analysis of genes. Nucleic Acids Res. 2022;50(W1):W782–90. https://doi.org/10.1093/nar/gkac392 .
Derkinderen P, Rolli-Derkinderen M, Chapelet G, Neunlist M, and Wendy Noble. Tau in the gut, does it really Matter? J Neurochem. 2021;158(2):94–104.
Doshi-Velez, Finale Y, Ge, and Isaac Kohane. Comorbidity clusters in Autism Spectrum disorders: an Electronic Health Record Time-Series Analysis. Pediatrics. 2014;133(1):e54–63.
Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–55.
Fuchs G, Voichek Y, Benjamin S, Gilad S, Amit I, and Moshe Oren. 4sUDRB-Seq: measuring Genomewide Transcriptional Elongation Rates and initiation frequencies within cells. Genome Biol. 2014;15(5):R69.
Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Milind Mahajan, et al. Most genetic risk for Autism resides with common variation. Nat Genet. 2014;46(8):881–85.
Grove J, Ripke S, Als TD, Manuel Mattheisen RK, Walters H, Won J Pallesen, et al. Identification of Common Genetic Risk variants for Autism Spectrum Disorder. Nat Genet. 2019;51(3):431–44.
Gutowska-Ding M, Weronika ZC, Deans C, Roos J, Matilainen F, Khawaja K, Brügger JW, Ahn C, Boustred, and Simon J. Patton. One byte at a time: evidencing the quality of clinical service next-generation sequencing for germline and somatic variants. Eur J Hum Genetics: EJHG. 2020;28(2):202–12.
Article PubMed Google Scholar
Heikkilä TE, Emilia K, Kaiser J, Lin D, Gill JJ, Koskenniemi. and Ville Karhunen. 2024. Genetic Evidence for Efficacy of Targeting IL-2, IL-6 and TYK2 Signalling in the Prevention of Type 1 Diabetes: A Mendelian Randomisation Study. Diabetologia, September. https://doi.org/10.1007/s00125-024-06267-5
Hemani G, Tilling K, George Davey Smith. Orienting the causal relationship between Imprecisely measured traits using GWAS Summary Data. PLoS Genet. 2017;13(11):e1007081.
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. https://doi.org/10.7554/eLife.34408 . PMID: 29846171; PMCID: PMC5976434.
Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, Joyce Y, Tung DA, Hinds RH, Perlis, and Ashley R. Winslow. Identification of 15 genetic loci Associated with risk of Major Depression in individuals of European descent. Nat Genet. 2016;48(9):1031–36.
Iwata K, Matsuzaki H, Tachibana T, Ohno K, Yoshimura S, Takamura H, Yamada K, et al. N-Ethylmaleimide-sensitive factor interacts with the Serotonin Transporter and modulates its trafficking: implications for pathophysiology in Autism. Mol Autism. 2014;5:33.
Jankowsky JL, Patterson PH. The role of cytokines and growth factors in seizures and their sequelae. Prog Neurobiol. 2001;63(2):125–49.
Jansen AG, Gwen C, Dieleman PR, Jansen FC, Verhulst D, Posthuma, Tinca JC, Polderman. Psychiatric Polygenic Risk scores as Predictor for attention Deficit/Hyperactivity Disorder and Autism Spectrum Disorder in a clinical child and adolescent sample. Behav Genet. 2020;50(4):203–12.
Kerimov N, Hayhurst JD, Peikova K, Manning JR, Walter P, Kolberg L, Samoviča M et al. 2021. eQTL Catalogue: A Compendium of Uniformly Processed Human Gene Expression and Splicing QTLs. bioRxiv. https://doi.org/10.1101/2020.01.29.924266
Kern JK, Geier DA, Sykes LK, Geier MR, Deth RC. Are ASD and ADHD a Continuum? A comparison of pathophysiological similarities between the disorders. J Atten Disord. 2015;19(9):805–27.
Khachadourian V, Mahjani B, Sandin S, Kolevzon A, Buxbaum JD. Abraham Reichenberg, and Magdalena Janecka. 2023. Comorbidities in Autism Spectrum Disorder and their etiologies. Translational Psychiatry 13 (1): 71.
Kuleshov MV, Jones MR, Rouillard AD, Nicolas F, Fernandez Q, Duan Z, Wang S Koplev, et al. Enrichr: a Comprehensive Gene Set Enrichment Analysis web server 2016 Update. Nucleic Acids Res. 2016;44(W1):W90–97.
Kwong A, Boughton AP, Wang M, VandeHaar P, Boehnke M. Gonçalo Abecasis, and Hyun Min Kang. 2022. FIVEx: an interactive eQTL browser across Public Datasets. Bioinformatics 38 (2): 559–61.
Lake BB, Chen S, Sos BC, et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 2018;36(1):70–80. https://doi.org/10.1038/nbt.4038 .
Li H. 2013. Aligning sequence reads, clone sequences and Assembly contigs with BWA-MEM. http://arxiv.org/abs/1303.3997
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, R. and 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics. 2009;25(16):2078.
Lin J, Tabassum R, Ripatti S, Pirinen M. MetaPhat: detecting and decomposing Multivariate associations from Univariate Genome-Wide Association Statistics. Front Genet. 2020;11:431.
Lin J, Mars N, Fu Y, Ripatti P, Kiiskinen T, Tukiainen T, Ripatti S. and Matti Pirinen. Integration of Biomarker Polygenic Risk Score Improves Prediction of Coronary Heart Disease. JACC: Basic to Translational Science. 2023. https://doi.org/10.1016/j.jacbts.2023.07.006
Lutz SM, Voorhies K, Wu AC, Hokanson J, Vansteelandt S, and Christoph Lange. The influence of unmeasured confounding on the MR Steiger Approach. Genet Epidemiol. 2022;46(2):139–41.
MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, et al. The New NHGRI-EBI Catalog of Published Genome-Wide Association Studies (GWAS catalog). Nucleic Acids Res. 2017;45(D1):D896–901.
Mattheisen M, Grove J, Als TD, Martin J, Voloudakis G, Meier S, Demontis D, et al. Identification of Shared and differentiating Genetic Architecture for Autism Spectrum Disorder, attention-deficit hyperactivity disorder and case subgroups. Nat Genet. 2022a;54(10):1470–78.
Mattheisen M, Grove J, Als TD, Martin J, Voloudakis G, Meier S, Demontis D, Bendl J, Walters R, Carey CE, Rosengren A, Strom NI, Hauberg ME, Zeng B, Hoffman G, Zhang W, Bybjerg-Grauholm J, Bækvad-Hansen M, Agerbo E, Cormand B, Nordentoft M, Werge T, Mors O, Hougaard DM, Buxbaum JD, Faraone SV, Franke B, Dalsgaard S, Mortensen PB, Robinson EB, Roussos P, Neale BM, Daly MJ, Børglum AD. Identification of shared and differentiating genetic architecture for autism spectrum disorder, attention-deficit hyperactivity disorder and case subgroups. Nat Genet. 2022b;54(10):1470–8. https://doi.org/10.1038/s41588-022-01171-3 . Epub 2022 Sep 26. PMID: 36163277; PMCID: PMC10848300.
Mazitov T, Bregin A, Philips M-A, Innos Jürgen, and Eero Vasar. Deficit in emotional learning in Neurotrimin Knockout Mice. Behav Brain Res. 2017;317(January):311–18.
McLaren W, Gil L, Hunt SE, Riat HS, Graham RS, Ritchie. Anja Thormann, Paul Flicek, and Fiona Cunningham. 2016. The Ensembl variant effect predictor. Genome Biol 17 (1): 1–14.
Morton JT, Dong-Min Jin RH, Mills Y, Shao G, Rahman D, McDonald Q, Zhu, et al. Multi-level analysis of the gut-brain Axis shows Autism Spectrum Disorder-Associated Molecular and Microbial profiles. Nat Neurosci. 2023;26(7):1208–17.
Nayar K, Sealock JM, Maltman N, Bush L, Cook EH, Lea K, Davis, and Molly Losh. Elevated polygenic burden for Autism Spectrum disorder is Associated with the broad autism phenotype in mothers of individuals with Autism Spectrum Disorder. Biol Psychiatry. 2021;89(5):476–85.
Nesterova AP, Yuryev A, Klimov EA, Zharkova M, Shkrob M, Ivanikova NV. Sergey Sozin, and Vladimir Sobolev. 2019. Disease pathways: an Atlas of Human Disease Signaling pathways . Elsevier.
Okbay A, Wu Y, Wang N, Jayashankar H, Bennett M, Nehzati SM, Sidorenko J, et al. Polygenic prediction of Educational Attainment within and between families from Genome-Wide Association Analyses in 3 million individuals. Nat Genet. 2022;54(4):437–49.
Palmer TM, Debbie A, Lawlor RM, Harbord NA, Sheehan JH, Tobias, Nicholas J, Timpson GD, Smith, Jonathan AC, Sterne. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21(3):223–42.
Pedersen CB, Bybjerg-Grauholm J, Pedersen MG, Grove J, Agerbo E, Bækvad-Hansen M, Poulsen JB, et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and Environmental Architectures of severe Mental disorders. Mol Psychiatry. 2018;23(1):6–14.
Peyre H, Schoeler T, Liu C, Michèle C, Williams N, Hoertel A, Havdahl, Jean-Baptiste P. Combining Multivariate genomic approaches to elucidate the comorbidity between Autism Spectrum disorder and attention deficit hyperactivity disorder. J Child Psychol Psychiatry Allied Discip. 2021;62(11):1285–96.
Phillips AN, Smith GD. How independent are ‘Independent’ effects? Relative risk estimation when correlated exposures are measured imprecisely. J Clin Epidemiol. 1991;44(11):1223–31.
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, et al. Convergence of genes and Cellular pathways Dysregulated in Autism Spectrum disorders. Am J Hum Genet. 2014;94(5):677–94.
Piovesan A, Caracausi M, Antonaros F, Pelleri MC, Vitale L. GeneBase 1.1: A Tool to Summarize Data from NCBI Gene Datasets and Its Application to an Update of Human Gene Statistics. Database: The Journal of Biological Databases and Curation 2016. 2016. https://doi.org/10.1093/database/baw153
Poplin R, Chang P-C, Alexander D, Schwartz S, Colthurst T, Ku A, Newburger D, et al. A Universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol. 2018;36(10):983–87.
Rees JMB, Angela M, Wood, and Stephen Burgess. Extending the MR-Egger Method for Multivariable mendelian randomization to correct for both measured and unmeasured pleiotropy. Stat Med. 2017;36(29):4705–18.
Risch N, Merikangas K. The future of genetic studies of Complex Human diseases. Science. 1996;273(5281):1516–17.
Romero M, Aguilar JM, Del-Rey-Mejías Ángel, Mayoral Fermín, Rapado Marta, Peciña Marta, Barbancho Miguel Ángel, Ruiz-Veguilla Miguel, Lara José Pablo. Psychiatric comorbidities in autism spectrum disorder: a comparative study between DSM-IV-TR and DSM-5 diagnosis. Int J Clin Health Psychol. 2016;16(3):266–75.
Ruotsalainen SE, Juulia J, Partanen A, Cichonska J, Lin C, Benner I, Surakka FG, et al. An expanded analysis Framework for Multivariate GWAS connects inflammatory biomarkers to functional variants and Disease. Eur J Hum Genetics: EJHG. 2021;29(2):309–24.
Sadik A, Dardani C, Pagoni P, Havdahl A, Stergiakouli E, iPSYCH Autism Spectrum Disorder Working Group, Khandaker GM, et al. Parental inflammatory bowel Disease and Autism in Children. Nat Med. 2022;28(7):1406–11.
Safran M, Dalah I, Alexander J, Rosen N, Stein TI, Shmoish M, et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010:baq020.
Schaaf CP, Betancur C, Yuen RKC, Jeremy R, Parr DH, Skuse L, Gallagher RA, Bernier, et al. A Framework for an evidence-based gene list relevant to Autism Spectrum Disorder. Nat Rev Genet. 2020;21(6):367–76.
Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6(2):461–64.
Singh K, Jayaram M, Kaare M, Leidmaa E, Jagomäe T, Heinla I, Hickey MA, et al. Neural cell adhesion Molecule Negr1 Deficiency in Mouse results in structural brain endophenotypes and behavioral deviations related to Psychiatric disorders. Sci Rep. 2019;9(1):5457.
Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, et al. Strengthening the reporting of Observational studies in Epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375:n2233.
Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen Sven Cichon, et al. Psychiatric genomics: an update and an agenda. Am J Psychiatry. 2018;175(1):15–27.
Suzuki H, Yoshida T, Morisada N, Uehara T, Kosaki K, Sato K, Matsubara K, Takano-Shimizu T, and Toshiki Takenouchi. De Novo NSF mutations cause early infantile epileptic Encephalopathy. Ann Clin Transl Neurol. 2019;6(11):2334–39.
Tamlander M, Mars N, Pirinen M, FinnGen E, Widén, and Samuli Ripatti. Integration of Questionnaire-based risk factors improves polygenic risk scores for human coronary heart disease and type 2 diabetes. Commun Biology. 2022;5(1):158.
Article CAS Google Scholar
Tamm L, Duncan A, Vaughn A, McDade R, Estell N, Birnschein A, and Lori Crosby. Academic needs in Middle School: perspectives of parents and youth with autism. J Autism Dev Disord. 2020;50(9):3126–39.
Tesi N, van der Lee S, Hulsman M, Holstege H, Marcel JT, Reinders. snpXplorer: a web application to explore human SNP-Associations and annotate SNP-Sets. Nucleic Acids Res. 2021;49(W1):W603–12.
Troisi J, Autio R, Beopoulos T, Bravaccio C, Carraturo F, Corrivetti G, Cunningham S, et al. Genome, Environment, Microbiome and Metabolome in Autism (GEMMA) Study Design: biomarkers identification for Precision Treatment and Primary Prevention of Autism Spectrum disorders by an Integrated Multi-omics Systems Biology Approach. Brain Sci. 2020;10(10). https://doi.org/10.3390/brainsci10100743 .
Turley P, Walters RK, Maghzian O, Okbay A, Mark Lee JJ, Fontana Alan, Nguyen-Viet Tuan Anh, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50(2):229–37.
Van den Hove DLA, Kenis G, Brass A, Opstelten R, Rutten BPF, Bruschettini M, Blanco CE, Lesch KP, Steinbusch HWM, Prickaerts J. Vulnerability versus resilience to prenatal stress in male and female rats; implications from Gene expression profiles in the Hippocampus and Frontal Cortex. Eur Neuropsychopharmacology: J Eur Coll Neuropsychopharmacol. 2013;23(10):1226–46.
Vgontzas A, and William Renthal. Migraine-Associated Gene expression in cell types of the Central and Peripheral Nervous System. Cephalalgia: Int J Headache. 2020;40(5):517–23.
Wallace C. Eliciting priors and relaxing the single causal variant Assumption in Colocalisation analyses. PLoS Genet. 2020;16(4):e1008720.
Wallace C. A more Accurate Method for Colocalisation Analysis allowing for multiple causal variants. PLoS Genet. 2021;17(9):e1009440.
Xie M-J, Iwata K, Ishikawa Y, Nomura Y, Tani T, Murata K, Fukazawa Y, and Hideo Matsuzaki. Autistic-like Behavior and Impairment of Serotonin Transporter and AMPA receptor trafficking in N-Ethylmaleimide sensitive factor gene-deficient mice. Front Genet. 2021;12:748627.
Download references
Acknowledgements
The work was supported by the European Commission Horizon 2020 programme (grant no. 825033) and the Strategic profiling of Tampere University in health data science, Academy of Finland, PROFI 6, 2021-2026. We are sincerely grateful to the entire GEMMA team and in particular to the nurses, recruiters and especially families participating in the project.
Open access funding provided by Tampere University (including Tampere University Hospital). The work was supported by the GEMMA project, funded by the European Commission by means of the Horizon 2020 program (call H2020-SC1-BHC-03-2018) with the project ID 825033 and the Strategic profiling of Tampere University in health data science, Academy of Finland, PROFI 6, 2021–2026.

Author information
Karoliina Salenius, Niina Väljä, Reija Autio and Jake Lin contributed equally to this work.
Authors and Affiliations
Faculty of Medicine and Health Technology, Tampere University and Tays Cancer Centre, Tampere, Finland
Karoliina Salenius, Niina Väljä, Sini Thusberg, Matti Nykter & Jake Lin
BMSystems, Paris, France
Francois Iris
Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, USA
Christine Ladd-Acosta
Euformatics, Tekniikantie, Espoo, Finland
Christophe Roos
Foundation for the Finnish Cancer Institute, Helsinki, Finland
Matti Nykter
European Biomedical Research Institute of Salerno (EBRIS), Salerno, Italy
Alessio Fasano
Harvard Medical School, Harvard T.H. Chan School of Public Health, Boston, USA
Health Sciences, Faculty of Social Sciences, Tampere University, Tampere, Finland
Reija Autio
Department of Medical Epidemiology and Biostatistics, Karolinska Institute, Stockholm, Sweden
You can also search for this author in PubMed Google Scholar
Contributions
JL, KS, CLA, CR, MN and RA designed the study. NV, KS, ST, RA and JL performed the data analysis and integration. KS, NV, ST, FI, RA and JL conducted the validation and figure generation. KS, FI, NV, AF, CR, RA and JL wrote the paper. All authors contributed to critical revisions and approved the final manuscript.
Corresponding author
Correspondence to Jake Lin .
Ethics declarations
Ethics approval and consent to participate.
The GEMMA study was approved by the relevant ethics committee of each enrolling country. Particularly, CE Campania Sud (IRB n.30/2019) for Italy; Partners Human Research (IRB ver.01/04/2019) for USA; and Clinical Research Ethics Committee of Galway University Hospital (IRB n. C.A. 2127/19) for Ireland. A written consent form will be signed by each participant or their legal representative.
Consent for publication
Consent, relevant to GEMMA subjects, is granted and signed by each participant or their legal representative.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary material 1., supplementary material 2., 12888_2024_6392_moesm3_esm.pdf.
Supplementary Figure 1. Genetic correlation of the traits included in the analysis. ASD = Autism Spectrum Disorder; ADHD = Attention Deficit Hyper Disorder; ASRD = Anxiety-Stress Disorder; DBD = Disruptive Behaviour Disorder; EA = Education attainment; MDD = Major Depression Disorder; SCZ = Schizophrenia. Supplementary Figure 2 . P -value and BIC decomposition processing of MAPT and NSF to identify ASD central traits. ASD=Autism Spectrum Disorder; ADHD=Attention Deficit Hyper Disorder; ASRD=Anxiety-Stress Disorder; DBD=Disruptive Behaviour Disorder; EA=Education attainment; MDD=Major Depression Disorder; SCZ=Schizophrenia. Supplementary Figure 3. Pathway analysis using the WikiPathway database also highlights neuronal processes, with bar length and color indicating significance. More details listed in Supplementary Table 8. Supplementary Figure 4. Tissue and cell (TS) type enrichment using WebCSEA and the list of the 22 central trait genes found that the most enriched tissues are related to cerebrum, cortex and small intestine related tissue types. Lake 2017 refers to data from human brain single cell analysis project (https://pubmed.ncbi.nlm.nih.gov/29227469/) while HCA stands for histologic chorioamnionitis, an intrauterine inflammatory trait. Supplementary Figure 5. ASD multivariate GWAS associations within the MAPT H1/H2 haplotype, 17q21 arm region, are presented in a Manhattan plot, in the context of Grove et al. GWAS results. Significance thresholds for p -values of 1e-05 indicated in blue and 1e-08 in red. Significant SNPs highlighted in green show rs62061734 (MAPT), rs269633 (KANSL1) and rs538628 (NSF).
12888_2024_6392_MOESM4_ESM.xlsx
Supplementary Table 1. Data and sample details of ASD and 8 genetically correlated traits ( P < 0.05, calculated from LD Score Regression (LDSC)) are presented and applied towards multivariate-GWAS. Data from four excluded traits are additionally shown. Supplementary Table 2. 37 multivariate associations are identified with ASD as a central trait where 17/37, shown with asterisk are previously reported in the GWAS Catalog and in bold, 8 genes are identified as SFARI ASD genes. Supplementary Table 3. 19 gene regions/trait pairings passed coloc (Posterior Prob H4 > 90%, Shown in bold) called on coloc.abf with a window size of ± 50 KB flanking the SNP locus. Supplementary Table 4. (A) Mendelian randomization (MR) results for ASD as outcome and related traits. (B) MR where ASD is the exposure and related traits are the outcome. Supplementary Table 5. MV associated genes are found in systems curated/implicated with gut microbiome and neural systems from GeneCards. Supplementary Table 6. List of 637 Significant SNPs ( p < 5e-8), with 315 already reported in the GWAS catalog, identified by MetaPhat multivariate-GWAS using ASD and 8 genetically correlated trait summary statistics. Supplementary Table 7. A) 108 enriched ( p < 0.05) Go terms are annotated and (B) 46 pathways on WikiPathway C) KEGG D) Reactome resources e) Tissue from the list of multivariate ASD SNPs found enrichments in neuron and nervous systems related data. Supplementary Table 8. ASD central SNP alleles are mapped to GEMMA genotypes called from 112 (49% females) WGS samples (45 (42% females) ASD probands). Phi coefficients are calculated between allele proportions where Chi-square test is applied to assess statistical importance. Indicated with *. Fisher's exact test is applied when Chi-square assumptions are violated. Supplementary Table 9. eQTL and sQTL related results of the ASD central associations relative to brain and nervous systems from EBI QTL catalog are captured via https://fivex.sph.umich.edu/. Study URLs are listed at the bottom of the table.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Salenius, K., Väljä, N., Thusberg, S. et al. Exploring autism spectrum disorder and co-occurring trait associations to elucidate multivariate genetic mechanisms and insights. BMC Psychiatry 24 , 934 (2024). https://doi.org/10.1186/s12888-024-06392-w
Download citation
Received : 01 August 2024
Accepted : 08 December 2024
Published : 18 December 2024
DOI : https://doi.org/10.1186/s12888-024-06392-w
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- ASD genetically correlated traits
- Multivariate GWAS
- Mendelian randomization
BMC Psychiatry
ISSN: 1471-244X
- General enquiries: [email protected]
An official website of the United States government
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock Locked padlock icon ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
- Publications
- Account settings
- Advanced Search
- Journal List

Autism Spectrum Disorder: Brain Areas Involved, Neurobiological Mechanisms, Diagnoses and Therapies
Jacopo lamanna, jacopo meldolesi.
- Author information
- Article notes
- Copyright and License information
Correspondence: [email protected]
Received 2023 Dec 5; Revised 2024 Jan 31; Accepted 2024 Feb 13; Collection date 2024 Feb.
Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( https://creativecommons.org/licenses/by/4.0/ ).
Autism spectrum disorder (ASD), affecting over 2% of the pre-school children population, includes an important fraction of the conditions accounting for the heterogeneity of autism. The disease was discovered 75 years ago, and the present review, based on critical evaluations of the recognized ASD studies from the beginning of 1990, has been further developed by the comparative analyses of the research and clinical reports, which have grown progressively in recent years up to late 2023. The tools necessary for the identification of the ASD disease and its related clinical pathologies are genetic and epigenetic mutations affected by the specific interaction with transcription factors and chromatin remodeling processes occurring within specific complexes of brain neurons. Most often, the ensuing effects induce the inhibition/excitation of synaptic structures sustained primarily, at dendritic fibers, by alterations of flat and spine response sites. These effects are relevant because synapses, established by specific interactions of neurons with glial cells, operate as early and key targets of ASD. The pathology of children is often suspected by parents and communities and then confirmed by ensuing experiences. The final diagnoses of children and mature patients are then completed by the combination of neuropsychological (cognitive) tests and electro-/magneto-encephalography studies developed in specialized centers. ASD comorbidities, induced by processes such as anxieties, depressions, hyperactivities, and sleep defects, interact with and reinforce other brain diseases, especially schizophrenia. Advanced therapies, prescribed to children and adult patients for the control of ASD symptoms and disease, are based on the combination of well-known brain drugs with classical tools of neurologic and psychiatric practice. Overall, this review reports and discusses the advanced knowledge about the biological and medical properties of ASD.
Keywords: arborization, dendrite, dysgenesis, flat, haploinsufficiency, organoid, pre-/post-synapse, pre-school, spine, subtype
1. Generalities about ASD
Autism Spectrum Disorder, ASD, is a neurodevelopmental brain complex disease established in growing fetuses and very young children during early development. ASD is caused by the altered expression of specific genes [ 1 , 2 ]. Its identification was not an early discovery. In 1943, the comprehensive brain disease of origin, autism, had been recognized in very young, pre-school children affected by biomedical-to-psychological problems accompanied by problems of communication and social interaction [ 3 , 4 , 5 ]. During the following three decades, the study of autism in children continued. Their neurons were recognized to express chromosomal defects including perturbed genes revealed during their structure organization. In addition, ASD was found to be very complex especially from its pathogenic point of view. Moreover, its factors, involved in various effects, were very heterogeneous. In 1993, distinct components of autism disorder began to be proposed and then wishfully recognized. ASD appears as the most significant disease in the field. Initially, its definition was widely debated; now, it is generally accepted as the complex neurodevelopmental disorder established during, and caused by, genetic and environmental perturbations [ 1 , 2 ].
In ASD patients, however, the affected genes are not always the same. Rather, they are complicated by its extensive heterogeneity. In other words, each ASD gene is expressed by a fraction, neither by all affected patients nor by the recognized animal models [ 3 , 4 , 5 ]. This increased understanding made possible the inclusion in ASD of a number of subcategories, which is important for their distinct clinical and therapeutic properties [ 4 ]. However, all attempts made to identify various heterogeneous genes within clear subtypes and subcategories resulted in limited successes [ 4 , 6 ].
In the brain of children, the ASD distribution in the gray matter is wide, including structures such as the midbrain, pontine, bilateral hippocampus, left para-hippocampal gyrus, various temporal and occipital gyruses. Together with their affected genes, these structures sustain deficits of communication and restricted (or repetitive) behavior. In addition to their altered neurogenic effects, ASD was co-affected by abnormalities. Some properties of the latter, however, are still largely uncharacterized [ 1 , 2 ]. The principal targets of the disease are the synapses, in particular the post-synapses of dendrites, both flat and spiny ( Figure 1 ). In addition to their general brain mentioned here, children with ASD show other, less frequent symptoms, including gastrointestinal dysfunctions and pulmonary hypertension. Organ alterations, such as scoliosis, are rare in children.
Examples of two types of dendrite arborization emerging from similar neuronal cell bodies and examples of dendritic spines. In ( A ), all dendritic fibers appear smooth because their post-synaptic structures, largely predominant in inhibitory neurons, are flat, i.e., they do not emerge or emerge only marginally from the fiber surface. In ( B ), the dendritic fibers predominant in stimulatory neurons are largely covered by spines, i.e., small stemming protrusions connected to fibers by their necks. ( A , B ) contain modified versions of Figure 1 from our previous publication [ 7 ]. ( C ) shows a fraction of an original figure by Santiago Ramon y Cajal (1896) (CAT 024, book Ciencia y Arte by the Instituto Cajal, Madrid, 2004) showing the heterogeneity of the spines emerging from dendritic fibers of pyramidal cells, illustrating in particular their variability in size, shape, density and distribution.
The child ASD frequency has been established in human populations. Up to 15 years ago, it was reported to account for 1.1% of the child population in the whole USA. As a consequence of recent intense investigations and advanced procedures employed, the calculated value corresponds to 2.3% of children. In a wider population, the affected children have been estimated to account for 1 out of 59 [ 1 ].
Given the extensive literature of ASD, it is vital to distinguish it from the other diseases. For this, in addition to the focus on child patients, it is important to illustrate various properties of the disease [ 8 ]. Therefore, the disease, including the aged form, has been defined also by psychodiagnostic tools in terms of DSM-5, i.e., according to the definition of Diagnostic and Statistical Manual for Mental Disorders from the American Psychiatric Association [ 9 ]. Prolonged treatments with factors have been analyzed also in mice [ 10 ]. By these definitions, we have emphasized the relevance of ASD in mental medicine, which is a concept that is re-discussed in the following Diagnosis Section 5 .
The frequency of ADS in adult patients has been found to be close to the frequency in children just mentioned (2.2%) [ 1 ]. Consequences of aging are structural alterations often resulting from the progressive degenerations of abnormal spinal curvature due to the progressive alteration of its elements [ 11 , 12 , 13 , 14 ]. These conditions often require minimally invasive surgery of the spinal curvature taking place upon the accurate preparation and recognition of risk factors: pain, loss of function, fatigue and many others [ 11 , 14 ]. In a minor number of these patients, surgeries need to be repeated [ 12 , 13 ]. In other adult patients, ASD lesions take place in the heart, affected by congenital atrial septal defects, and others take place in the bones [ 15 , 16 ]. Both these lesions are often treated by repetitive surgery [ 16 ]. The ASD pathology of children and adults is therefore largely different.
The present review is based on two main origins: established articles published one or two decades ago, reporting key documents about the existence and relevance of ASD, necessary for a recognized disease, followed by more recent publications including those that appeared during the last year. Within these two origins, many publications have been selected by direct comparison with others, which are similar in properties but less complete and precisely focused. Similar criteria have been used in the choice of ASD with respect to other subcategories, such as Asperger’s syndrome [ 17 , 18 ], which is also generated, but distinct, in the autism area. These other subcategories are not presented in parallel to ASD but rather only mentioned. General definitions such as intellectual disability (ID) and a few others, employed in previous publications for the completion of ASD properties [ 2 ], are not presented here in detail. In other words, our intention has been to inform our readers about ASD, including knowledge established recently.
2. The ASD Structures in the Brain
ASD children begin to be investigated during the first months of life, when the brain volume appears normal. During the subsequent months, the volume grows, reaching increases of approximately 10% at ages between 2 and 4 years. During late childhood and adolescence, the brain volume increases further but more slowly. Interestingly, the increased volume of many areas is more evident in the left than in the right brain. In particular, increases are reported in the hippocampal and temporal roles, in which however gray matter tends to decrease and white matter tends to increase [ 19 ]. Decreases of another structure, the cerebellum, are appreciable especially in adolescent and adult patients [ 20 ]. The details of structural properties, reported in many studies, have been investigated in populations of ASD patients. The considerable heterogeneity of the disease corresponds in many cases to functional alterations typical of various ASD subtypes [ 1 , 2 , 4 , 19 , 20 ].
Knowledge and understanding of the changes reported in ASD are critical to establish appropriately the state of neural structures in living patients. Adequate results for this purpose have been obtained by magnetic resonance imaging (MRI), which was initially employed more than a decade ago [ 21 ]. Recently, MRI has been employed in its multimodal forms, which are appropriate for early clinical diagnoses and also for present perspectives of therapy [ 22 , 23 , 24 ].
The changes of brain properties reported in ASD patients are largely due to their size and distribution. In children, the brain volume changes are primarily due to the increased surface area rather than cortical thickness [ 24 , 25 ]. White matter development can occur [ 25 ]; however, in several areas, the alterations are mostly due to gray matter. In the amygdala and hippocampus, the changes of gray matter, associated with the severity of autistic symptoms and language, are often reduced [ 26 , 27 ]. Other changes of ASD’s local properties take place during patient aging. For example, cortical gyrification is increased in children but decreases rapidly upon the development of adolescence [ 28 ].
3. Mechanisms of ASD Gene Operations: Key Role of Synapses
Gene variants of ASD, together with their regulatory nuclear factors often called risks, induce marked changes in a wide range of biological processes. The identification of some gene mutants has confirmed the ASD risks acting through largely distinct molecular pathways. Thus, the genes of ASD and the ensuing epigenetics are finely modulated in their human genomic context [ 29 , 30 ]. With time, the association of gene variants with ASD risks has been investigated, and many positive cases have been reported [ 31 , 32 , 33 ]. In some cases, however, the relevance of such variants has remained unclear. In order to solve the problems, some questions have been characterized in terms of nucleotide polymorphisms. The results have demonstrated the relevance of some genes and not other genes related to ASD [ 34 ]. Analogously unidentified ASD genes govern neuronal structure–function relationships in the cortex and other brain areas [ 35 ]. Moreover, a gene mutant, Sparcl1, and its protein Hevin, active as an ASD risk, have been found to induce, in the endoplasmic reticulum of neurons, a sort of stress characterized by structural instability [ 36 ]. The growing architecture induced by genetic variants is now providing innovative information about the pathological role of human ASD [ 32 , 37 ].
In addition to genes, two other factors are essential for ASD function: transcription factors (for example [ 38 ]) and the protein complexes active on chromatin remodeling. A very active subunit of the latter type, reported first in 2016 [ 39 ], is ARID1B. The latter complex, by interacting with various genes, plays a primary role in the growth of neurons and thus in ASD of children and adolescents [ 40 ]. Its knock-down was found to induce a decreased arborization of dendrites with an ensuing decrease in both excitatory and inhibitory intercellular communications [ 39 , 41 ].
Additional studies confirmed that marked changes of gene expression, operative with ARID1B, take place together with the decreased body size of the brain. The basic alterations include cortical inhibitory/excitatory imbalance with decreased GABAergic neurons and their transmission [ 41 ]. Interestingly, haploinsufficient mice are protected by the administration of growth hormone and also by early postnatal serotonin modulation [ 42 ]. ARID1B, therefore, appears to play an essential role in forebrain neurogenesis, inducing a pronounced role in inhibitory neural progenitors [ 40 , 41 , 42 ], with refinement of the ensuing progressive therapy [ 43 ]. In addition, available evidence has demonstrated that the role of ARID1B is not unique. Analogous effects are in fact induced by other chromatin remodeling subunits. This has been the case of KANSL1, WDR5 and a few others, which have been integrated into the present knowledge of ASD pathogenesis [ 2 ].
Within neurons, the structures and fractions most widely affected by ASD genes and proteins correspond to, and interact with, various types and components of synapses [ 44 ]. In addition to intellectual and social effects, disabilities and speech defects, the impairment mutations of chromatin remodeling induced by ARID1B regulate primarily the dendritic differentiation in the developing brain [ 43 ]. The dysfunction of dendrites, the post-synaptic fibers that receive the pre-synaptic inputs, are critical for synaptic function, and they are also relevant in sensory processing, cognition and conscious perception [ 44 ]. The knock-down of ARID1B results in a block of stimulatory synaptic transmission ( Figure 2 ).
ARID1B acts as a scaffolding protein that holds together the components of its complex ability to operate with specific chromatin components of reactive genes. ( A , C ) images illustrate two cells (orange) characterized by flat and spiny dendritic fibers, respectively. In the nucleus (violet) of these images, the ARID1B complex regulates the transcription of specific genes. The generated mRNA transcripts (small black dots) are transferred to the cytoplasm of the corresponding proteins addressed to the dendritic fibers (red arrows). In ( A ), the latter proteins contribute to the appropriate assembly of flat post-synaptic structures. The bottom ( B ) is analogous to ( A ) except that ARID1B has been knocked-down, the red pointed arrows do not move specific mRNAs, small white dots contain proteins different from those generated by ARID1B, the post-synapses are absent, and the pre-synapses are scattered in the space. ( C ) is like ( A ) except for one spine with black dots assembled close to two pre-synapses assembling whole synapses; ( D ) corresponds to ( C ) without ARID1B; thus, it is analogous to ( B ) with respect to ( A ). The change in ( D ) versus ( C ) is the tiny spine to which pre-synapses assemble to establish the whole synapse.
Moreover, the integration of pre-synapses with dendritic excitability is reduced together with the number and morphology of dendritic spines. The dendrites with their post-synaptic responses to flat and spines are therefore sites of ASD gene effects [ 2 , 39 , 42 , 44 ]. In addition to the molecular complexes active as chromatin modifiers [ 42 , 43 , 44 ], ARID1B and its analogous subunits have been shown to govern other genes, encoding proteins localized at or near synapses [ 43 , 44 , 45 ] ( Figure 2 ).
Specific aspects of synapses affected by ASD action need to be considered in structural and functional terms. In ASD pre-synapses, the structure and release of neurosecretory generation are less frequently affected [ 41 , 42 ], except for small boutons and glutamatergic synapses, whose post-synaptic mGluR is governed by Shank3 mutations and the interaction with neuroligin 1 [ 44 , 45 , 46 ]. On the other hand, the main structures of post-synapses, dendrites with their flat and spine structures, predominant in inhibitory and stimulatory synapses, respectively, are critical also because of their key role in the transmission from pre-synaptic structures [ 41 , 46 ]. The spines, tiny post-synaptic protrusions from dendrites that receive most of the excitatory synaptic input, are almost always affected by their role in ASD ( Figure 1 and Figure 2 ) [ 2 , 42 , 46 , 47 , 48 ]. The predominant role of spines has been confirmed by studies where various aspects of synaptic ASD are investigated in parallel [ 43 , 44 ]. Functional and structural changes of spines are critical for synaptic plasticity, which is a cellular model of learning and memory. Altered spine morphology and plasticity are common markers of human neurodevelopmental disorders, such as conformational fluctuations and ID [ 49 , 50 , 51 , 52 ].
A final problem, not yet discussed in this review, deals with additional processes that activate ASD functions. In a mouse model, a dramatic increase in NO level induces a marked worsening of the ASD state. Moreover, high levels of nitrosative stress biomarkers and NO synthase inhibitors were found to induce changes of behavioral ASD-associated phenotype [ 53 ]. Analogous increases of oxidative stress were found to induce a polymorphism of the NRF2 gene, which is a master regulator of antioxidant stress [ 54 ]. Additional known stresses were found effective in the expression of ASD genes [ 55 , 56 ]. Interesting results were also obtained with human pluripotent stem cells assembled in organoid models [ 30 ]. These models, generated through the use of human pluripotent stem cells, produce profoundly intricate systems with spatiotemporal modeling of the developing brain by approaches that appear promising; however, they are still limited [ 57 ]. Based on previous suggestions and the present models, we can conclude that in the near future, some innovative ASD aspects will be recognized and characterized.
4. Contribution of Glial Cells
Up to now, the presentation of ASD and its regulation have been considered only of a neuronal nature. High numbers of glial cells, however, are present in the brain and at least two of them, astrocytes and microglia, discussed extensively in the literature during the last few years, participate in critical aspects of ASD. Here, we summarize a few relevant aspects of the glial cell role in ASD. Results of the two types of cells are reported, first concerning their separate effects [ 58 , 59 , 60 , 61 , 62 ] and then working together with coordinate effects [ 63 , 64 ].
In a normal brain, the number of astrocytes is considerable. In ASD-positive brains, such a number is decreased; however, the remaining astrocytes are often active. It can be concluded that astrocytes play a protective role on neuronal functions [ 59 ] with ensuing changes of synapse function [ 59 ]. The astrocyte roles, including brain inflammation, participate directly in the functions sustained by neurons [ 59 , 60 ].
The ASD role of the second type of glial cells Is focused more precisely. Microglia is strongly active in inflammation. Therefore, it does increase the process substantially. In numerous processes sustained by astrocytes together with neurons, microglia is inhibited. This occurs with neuroligin-4, which is a factor that interacts directly with ASD and operates also on other processes [ 61 ]. A factor that prevents inflammation is minocycline, which operates by modulating microglia polarization and therefore protecting ASD [ 62 ].
During the coordinate activations of the two glial cell types, some of their combined changes are relevant also for ASD. For example, microglia activation often induces astrocyte reactive activation and the ensuing release of ATP, which further activates microglia [ 63 ]. Moreover, chemokines and factors released by microglia are blocked by other factors released by astrocytes. The effects of both glial cells investigated in terms of ASD are shown in [ 64 ].
The most interesting interactions of glial cells include those with neurons. Specifically, accurate analyses of neuroinflammation, interactive with immunometabolic factors of glial origin, as well as immune mediators of ASD patients interactive with gene mutations, were reported to result in stress-positive responses [ 65 ]. The interest in these results was related to future therapies against immune abnormalities of children ASD [ 29 ]. On the other hand, ASD generation by growing fetuses was found to be reinforced by maternal antibodies; however, there was no convergence of stress functions [ 66 ]. Another important process, the generation of ASD markers, was shown to depend on miRNA expression by a serotonin transporter gene [ 67 ].
5. Diagnoses
ASD symptoms and pathological properties have already been mentioned previously. The goal of this section is not a repetition but rather the order presentation of advanced processes of diagnosis in relation to ASD alone and in combination/relation with other diseases. Diagnosing requires understanding how autistic patients react in response to various value-based paradigms [ 68 , 69 ]. This information should be considered in the early identification and diagnosis of ASD. At present, most family parents are concerned about their diseased children even before they turn 2 years old, even when their diagnoses are not made until age 4 or later. Under these conditions, the state of patients is often impaired due to their aggravating symptoms. Relevant especially for poor communities, recognition of the disease by experienced clinicians is essential in many terms: medical, operational and even economical [ 70 , 71 ]. For ASD children, diagnoses are often made by the application of advanced tests, such as the psychodiagnostic tests, a gold standard in diagnosing ASD, by including in the evaluations also the criteria of DSM-5 defined by the Diagnostic and Statistical Manual for Mental Disorders [ 9 , 72 ]. In case the results remain uncertain, additional distinct tests can be employed based on the experience of specialized centers [ 68 , 73 ].
In addition to children, analogous diagnostic efforts are made for adult ASD patients. For these symptoms, the efforts are numerous, including impaired social interactions, limited communication skills, and repetitive behaviors [ 9 , 69 ]. In addition, biochemical work-up can include body fluid analyses to reveal general metabolic and lysosomal storage properties; changes in autistic symptoms can result in self-injurious behaviors and psychomotor responses; genomic technology can be employed to identify molecular defects [ 32 , 33 , 74 , 75 ]. For patients of advanced age, appropriate diagnoses are very important also in term of destiny. Compared to the general population, in fact, their mortality is 2.9-fold higher [ 74 ].
Additional diagnoses of ASD are made based on specific properties we have already reported in previous sections. ARID1B haploinsufficiency is the well-known subunit complex effect of chromatin remodeling already presented [ 39 , 40 , 41 , 42 ]. Its related disorders, important for the recognition of ASD phenotypes, are highly heterogeneous. Animal models of this pathology have helped to identify the ASD molecular mechanisms where ARID1B participates in brain development [ 76 ]. In addition to molecular mechanisms, disease aspects are regulated by metabolic and cytoplasmic structures [ 33 ]. Convergent mechanisms underlying the dysgenesis of dendritic spines contribute to the distinction of ASD from other pathologies, which has been investigated in several animal models and in human post-mortem brain samples. Another concept developed by the study of ASD children, either alone or combined with another defect, is attention deficit hyperactive disorder (ADHD). The study confirmed ADS children expressing levels of anxiety higher than those of their peers [ 77 ]. They also exhibit higher levels when co-expressed with ADHD. Interestingly, results analogous to those of anxiety, induced by separate and combined ASD and ADHD, have been found also when ASD is combined with other processes such as gender dysphoria and impaired locomotor skills. Therefore, the results with child ASD together with ADHD can be interpreted as a form of psychiatric comorbidity [ 77 , 78 ].
Another example of ASD comorbidity is that of child sleep. The results demonstrated that sleep problems affect more than 95% of the patients and over 86% of their father parents. The latter 86% parents have more anxiety and depression than the parents of ASD children with no sleep problems. These results reveal how child sleep problems affect the well-being of parents [ 79 ]. Future research will establish whether comorbidities similar to those of sleep problems induce in parents also other effects, such as those induced in children by ASD [ 79 ].
The brain diseases and properties considered so far have been shown to depend directly on ASD. In other words, independent diseases co-occur with autistic defects. This state does not cover the whole types of ASD dependence. In the USA, when compared with the corresponding peers, the brain diseases expressed by ASD in children neurons induce depression frequencies of about 3-fold, anxiety of 2-fold, and epilepsy of over 20-fold [ 1 ]. Examples of corresponding therapy are reported in the following Section 7 . Analogous studies have been made in adult ASD patients for neurodegenerative diseases assayed in terms of genes, symptoms and effects involved. An overlap of ASD risk genes has been found with schizophrenia. With neurodegenerative diseases, in particular with Parkinson’s and Alzheimer’s diseases, some gene commonality has emerged [ 80 , 81 ]. At present, however, details remain to be confirmed.
6. Therapies
The goal of many ASD studies, in particular those about diagnosis (see for example [ 65 , 75 , 80 , 81 ]), has been dedicated to specific therapies, first based on the present knowledge and then translated into clinical use. A problem of this approach is the heterogeneity of analyzed patients, especially of small infants, which is particularly difficult to characterize. The solution of this problem is expected from learning machines associated with electroencephalography and/or magnetoencephalography [ 82 ].
Drug employment is the mainstay of treatment for the core symptoms of ASD, including communication deficits, social interaction deficits and repetitive behavior. For the last 10 years, risperidone and aripiprazole, officially two antipsychotic drugs, are the only ones approved by the FDA to treat ASD children’s irritability. At present, the two drugs are employed in more than 30 countries. The doses per patient used in the USA and Europe are much higher than those in Turkey and other eastern countries [ 1 , 83 ]. However, the effectiveness of these drugs is limited, and adverse effects are frequent. The attempts of the industry for the production of new drugs or the employment of drugs already on the market are intense; however, the results are limited. Effects of cannabinoids and interest for probiotics, prebiotics and symbiotics are promising [ 84 , 85 ]; however, these are not yet approved.
At present, the ongoing ASD therapy is limited to well-known brain drugs prescribed to both children and adult patients for many ASD co-symptoms: anxiety, depression, hyperactivity, and sleep problems [ 86 , 87 ]. Among these drugs, those effective against attention-deficit/hyperactivity disorders are employed as psychotropic and psychostimulants [ 1 , 86 , 87 , 88 ]. Ongoing research, now intensely investigated, supports an association between ASD and immune/inflammatory mechanisms and proposes the development and future employment for specific inhibitory drugs [ 89 ]. Improvements of cognitive rehabilitation have been obtained by changes in the eye movement’s performance [ 90 ]. The administration of some such drugs induces various tolerability effects, starting by weight increase. Most often, the negative effects are controlled by adjustment of the employed doses [ 86 , 87 , 88 , 89 ].
Disorders not strictly dependent on ASD, such as gastrointestinal disorders, are also employed for ongoing therapy. Recent studies have revealed the brain mechanisms of such drugs. The microbiota gut–brain axis operates as a modulator of neuropsychiatric health. Many important functions, such as brain cognitive actions, as well as immunities, are governed by gut metabolites [ 91 ]. In combination with drugs, treatments include non-pharmacological interventions, such as behavioral therapy and acupuncture. Behavioral therapies, largely provided by doctors to both children and adult patients, have been often evaluated favorably based on high or moderate evidence [ 5 , 92 ]. Acupuncture has been found to stimulate some social functioning; however, the positive evidence of this therapy is limited [ 92 ]. Summing up, many well-known brain drugs are employed in ASD therapy with appreciable but not innovative results. A recent review has reconsidered critically the list of most such drugs. The results have largely confirmed the previous data. In addition, two long-term drugs, amitriptyline and loxapine, have been promising, deserving specific trials not yet dedicated to them for ASD diseases [ 93 ].
7. Conclusions
As specified in the title, the main interest of this review is focusing on various aspects of ASD medicine investigated upon the identification of ASD during the last few years. In contrast, we have left out the pre-medical properties of patients, critical for the discovery of autism in 1943, concerning the deficits of language, communication and social interactions. Here, we do not provide specific presentations of the latter processes. In case of interest by readers, they can be found in various forms of the literature such as [ 94 , 95 ].
Upon the identification of ASD, specific studies have identified many but not all of the properties, such as those of synapses. The study of these properties will be continued during the next few years. We expect ensuing developments, especially from the dendrites and their post-synaptic structures, and then the involvement of various medical brain specialties: from biomedicine to neurology, psychiatry, and also brain surgery. At the moment, also in non-advanced countries, the cooperation of these specialties is sustaining social and political initiatives.
A highly important development reported in this review concerns the expected progress of ASD therapy. At present, together with a few of marginal relevance, the drugs employed for ADS are the same employed for the other brain diseases. Therefore, innovative drugs are needed, and the knowledge appears promising for future developments [ 96 , 97 ]. The emergence of pharmaceutical industrial initiatives appears to be even more innovative. If the numbers of appropriate polymers exposed to manufacturing technologies are limited, ongoing work is expected to develop new ASD drug formulations [ 98 ].
Abbreviations
Author contributions.
The two authors have developed the innovative improvements in the field and realized their presentation. The final version of this review has been written by J.M. and then critically analyzed together with J.L. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The author declares no conflicts of interest.
Funding Statement
This research received no external funding.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
- 1. Hirota T., King B.H. Autism spectrum disorder: A review. JAMA. 2023;329:157–168. doi: 10.1001/jama.2022.23661. [ DOI ] [ PubMed ] [ Google Scholar ]
- 2. Ford T.J.L., Jeon B.T., Lee H., Kim W.Y. Dendritic spine and synapse pathology in chromatin. Front. Mol. Neurosci. 2022;15:1048713. doi: 10.3389/fnmol.2022.1048713. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 3. Forstein S.E., Rosen-Sheidly B. Genetics of autism: Complex etiology for a heterogeneous disorder. Nat. Rev. Genet. 2001;2:943–955. doi: 10.1038/35103559. [ DOI ] [ PubMed ] [ Google Scholar ]
- 4. Fombonne E.J. Epidemiological surveys of autism and other pervasive developmental disorders: An update. Autism Dev. Disord. 2003;33:365–382. doi: 10.1023/A:1025054610557. [ DOI ] [ PubMed ] [ Google Scholar ]
- 5. Su Maw S., Haga C. Effectiveness of cognitive, developmental, and behavioral interventions for autism spectrum disorder in preschool-aged children: A systematic review and meta-analysis. Heliyon. 2018;4:e00763. doi: 10.1016/j.heliyon.2018.e00763. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 6. Angelink van Entergem J.A., Deserno M.K., Geurts H.M. Validation strategies for subtypes in psychiatry: A systematic review of research on autism spectrum disorder. Clin. Psychol. Rev. 2021;87:102033. doi: 10.1016/j.cpr.2021.102033. [ DOI ] [ PubMed ] [ Google Scholar ]
- 7. Meldolesi J. Dendritic post-synapses in the brain: Role of flat and spine structures. Biomedicines. 2022;10:185. doi: 10.3390/biomedicines10081859. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 8. Maurice V., Russet F., Scocco P., McNicholas F., Santosh P., Sing S.P., Street C., Purper-Ouakil D. Transition from child and adolescent mental health care to adult services for young people with attention-deficit/hyperactivity disorder (ADHD) or autism spectrum disorder (ADS) in Europe: Barriers and recommendations. Encephale. 2022;48:555–559. doi: 10.1016/j.encep.2022.01.012. [ DOI ] [ PubMed ] [ Google Scholar ]
- 9. Boccaccio F.M., Platania A.G., Savia Guerrera C., Varrasi S., Privitera R.C., Caponnetto P., Porrone C., Castellano S. Autism Spectrum Disorder: Recommended psychodiagnostic tools for early diagnosis. Health Psychol. Res. 2023;11:77357. doi: 10.52965/001c.77357. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 10. Park G., Jang W.E., Kim S., Gonzales E.L., Ji J., Choi S., Kim Y., Park J.-H., Mohammad H.B., Bang G., et al. Dysregulation of the Wnt/ β-catenin signaling pathway via Rnf146 up-regulation in a VPA-induced mouse model of autism spectrum disorder. Exp. Mol. Med. 2023;55:1783–1794. doi: 10.1038/s12276-023-01065-2. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 11. Ohyama S., Kotani T., Iijima Y., Okuwaki S., Sunami T., Iwata S., Sakuma T., Ogata Y., Akazawa T., Shiga Y., et al. Incidence and potential risk factors of superior mesenteric artery syndrome after spinal corrective surgery in patients with adult spinal deformity. World Neurosurg. 2023;5:e591–e598. doi: 10.1016/j.wneu.2023.09.114. [ DOI ] [ PubMed ] [ Google Scholar ]
- 12. Mir J.M., Galetta M.S., Tretiakov P., Dave P., Lafage V., Lafage R., Schoenfeld A.J., Passies P.G. Achievement and maintenance of optimal alignment following adult spinal deformity corrective surgery: A 5 years outcome analysis. World Neurosurg. 2023;180:e523–e527. doi: 10.1016/j.wneu.2023.09.106. [ DOI ] [ PubMed ] [ Google Scholar ]
- 13. Lee B.J., Bae S.S., Choi H.Y., Park J.H., Hyun S.J., Jo D.J., Cho Y. Proximal junctional kyphosis or failure after adult spinal deformity surgery-review of risk factor and its prevention. Neurospine. 2023;20:863–875. doi: 10.14245/ns.2346476.238. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 14. Dalton J., Mohammed A., Akioyamen N., Schwab F.J., Lafage V. Preoperative planning for adult spinal deformity goals: Level selection and alignment goals. Neurosurg. Clin. N. Am. 2023;34:527–536. doi: 10.1016/j.nec.2023.06.016. [ DOI ] [ PubMed ] [ Google Scholar ]
- 15. Elsayed Y.M.H., Almarghany A.A. Resolution of trifascicular heart block with effective closure of congenital atrial septal defect followed by later coronavirus disease 2019. J. Innov. Card. Rhythm. Manag. 2023;14:5533–5536. doi: 10.19102/icrm.2023.14081. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 16. Kawabata A., Sakai K., Yamada K., Utagawa K., Hashimoto J., Morishita S., Matsukura Y., Oyaizu T., Hirai T., Inose H., et al. The lower osteotomy level is associated with decreased revision surgery due to mechanical complications after tree column osteotomy in patients with adult spinal deformity. Glob. Spine J. 2023;18:21925682231196449. doi: 10.1177/21925682231196449. [ DOI ] [ PubMed ] [ Google Scholar ]
- 17. Faridi F., Seyedebrahimi A., Khorowabadi R. Brain structural covariance network in Asperger syndrome differs from those in autism spectrum disorder and healthy controls. Basic Clin. Neurosci. 2022;13:815–838. doi: 10.32598/bcn.2021.2262.1. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 18. Hosseini S.A., Molla M. Asperger Syndrome. StatPearls Publishing; Treasure Island, FL, USA: 2024. [ PubMed ] [ Google Scholar ]
- 19. Li C., Zhang R., Zhu Y., Li T., Quin R., Li L., Yuan X., Wang L., Wang X. Gray matter asymmetry alterations in children and adolescents with comorbid autism spectrum disorder and attention-deficit/hyperactivity disorder. Eur. Child Adolesc. Psychiatry. 2023:1–12. doi: 10.1007/s00787-023-02323-4. [ DOI ] [ PubMed ] [ Google Scholar ]
- 20. Kumar M., Hiremath C., Kumar Khokhar S., Bansai E., Vijay Sagar K.J., Patmanabha H., Grimaji A.S., Narayan S., Kishore M.T., Yamini B.K., et al. Altered cerebellar lobular volumes correlates with clinical deficits in siblings and children with ASD: Evidence from toddlers. J. Transl. Med. 2023;21:246. doi: 10.1186/s12967-023-04090-x. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 21. Schumann C.M., Bloss C.S., Barnes C.C., Wideman G.M., Carper R.A., Aksoomoff N., Pierce K., Hagler D., Schork N., Lord C., et al. Longitudinal magnetic resonance imaging study of cortical development through early childhood in autism. J. Neurosci. 2010;30:4419–4427. doi: 10.1523/JNEUROSCI.5714-09.2010. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 22. Akhavan A.M., Sharifi A., Pedram M.M. Combination of rs-fMRI and sMRI data to discriminate autism spectrum disorders in young children using deep belief network. J. Digit. Imaging. 2018;31:895–903. doi: 10.1007/s10278-018-0093-8. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 23. Hazlett H.C., Gu H., Munsell B.C., Kim S.H., Styner M., Wolff J.J., Elison J.D., Swanson M.R., Zhu H., Botteron K.N., et al. Early brain development in infants at high risk for autism spectrum disorder. Nature. 2017;542:348–351. doi: 10.1038/nature21369. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 24. Ohta H., Nordahl C.W., Iosif A.M., Lee A., Rogers S., Amaral D.G. Increased surface area, but not cortical thickness, in a subset of young boys with autism spectrum disorder. Autism Res. 2016;9:232–248. doi: 10.1002/aur.1520. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 25. Andrews D.S., Lee J.K., Harvey D.J., Waizbard-Bartov E., Solomon M., Rogers S.J., Nordhal C.W., Amaral D.J. A longitudinal study of white matter-development in relation to changes in autism severity across early childhood. Biol. Psychiatry. 2021;89:424–432. doi: 10.1016/j.biopsych.2020.10.013. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 26. Sun F., Chen Y., Gao Q., Zhao Z. Abnormal gray matter structure in children and adolescents with high-functioning autism spectrum disorder. Psychiatry Res. Neuroimaging. 2022;327:111564. doi: 10.1016/j.pscychresns.2022.111564. [ DOI ] [ PubMed ] [ Google Scholar ]
- 27. Arutiunian V., Davydova E., Pereverzeva D., Sorokin A., Tyushkevich S., Mamokhina U., Danilina K. Reduced grey matter volume of amygdala and hippocampus is associated with the severity of autistic symptoms and language abilities in school-aged children with autism spectrum disorder: An exploratory study. Brain Struct. Funct. 2023;228:1573–1579. doi: 10.1007/s00429-023-02660-9. [ DOI ] [ PubMed ] [ Google Scholar ]
- 28. Kohli J.S., Kinnear M.K., Fong C.H., Fishman I., Carper R.A., Muller R.A. Local cortical gyrification is increased in children with autism spectrum disorders, but decreases rapidly in adolescents. Cereb. Cortex. 2019;29:2412–2423. doi: 10.1093/cercor/bhy111. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 29. Paulsen B., Velasco S., Kedaigle A.J., Pigoni M., Quadrato G., Deo A.J., Adikonis X., Uzquiano A., Sartore R., Yang S.M., et al. Autism genes converge on asynchronous development of shared neuron classes. Nature. 2022;602:268–273. doi: 10.1038/s41586-021-04358-6. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 30. Lenon de Souza Santos J., Almeida Arauio C., Arauio Gurge Rocha C., Costa-Ferro Z.S., Solano de Freitas Souza B. Modeling autism spectrum disorders with induced pluripotent stem cell-derived brain organoids. Biomolecules. 2023;13:260. doi: 10.3390/biom13020260. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 31. Cai C., Yin Z., Liu A., Wang H., Zeng S., Wang Z., Qiu H., Li S., Zhu X., Wang M. Identifying rare genetic variants of immune mediators as risk factors for autism spectrum disorder. Genes. 2022;13:1098. doi: 10.3390/genes13061098. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 32. Lima Caldeira G., Peca J., Carvalho A.L. New insights on synaptic dysfunction in neuropsychiatric disorders. Curr. Opin. Neurobiol. 2019;57:62–70. doi: 10.1016/j.conb.2019.01.004. [ DOI ] [ PubMed ] [ Google Scholar ]
- 33. Senarathne U.D., Indika N.L.R., Izela-Sanek A., Ciara E., Frye R.E., Chen C., Stepien K.M. Biochemical, genetic and clinical diagnostic approaches to autism-associated inherited metabolic disorders. Genes. 2023;14:803. doi: 10.3390/genes14040803. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 34. Fang Y., Cui Y., Yin Z., Hou M., Guo P., Wang H., Liu N., Cai C., Wang M. Comprehensive systematic and meta-analysis of the association between common genetic variants and autism spectrum disorder. Gene. 2023;18:147723. doi: 10.1016/j.gene.2023.147723. [ DOI ] [ PubMed ] [ Google Scholar ]
- 35. Dong D., Li J., Ju Y., Xiao C., Li K., Shi B., Zheng W., Zhang Y. Altered relationship between functional connectivity and fiber bundle structure in high functioning male adults with autism spectrum disorder. Brain Sci. 2023;12:1098. doi: 10.3390/brainsci13071098. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 36. Taketomi T., Yasuda T., Morita R., Kim J., Shigeta Y., Eroglu C., Harada R., Tsuruta F. Autism associated mutations in Hevin/Sparcl1 induces endoplasmic reticulum stress through structural instability. Sci. Rep. 2022;12:11891. doi: 10.1038/s41598-022-15784-5. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 37. Nishioka M., Takayama J., Sakai N., Kazuno A.A., Ishiwata M., Ueda J., Hayama T., Fujji K., Samaya T., Kuriyama S., et al. Deep exome sequencing identifies enrichment of deleterious mosaic variants in neurodevelopmental disorder genes and mitochondrial tRNA regions in bipolar disorder. Mol. Psychiatry. 2023;28:4294–4306. doi: 10.1038/s41380-023-02096-x. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 38. Caracci M.O., Avila M.E., Espinosa-Cavieres F.A., Lopez H.R., Ugarte G.D., De Ferrari G.V. Wnt/βcatenin-dependent transcription in autism spectrum disorder. Front. Mol. Neurosci. 2021;14:764756. doi: 10.3389/fnmol.2021.764756. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 39. Ka M., Chopra D.A., Dravid S.M., Kim W.Y. Essential roles for ARID1B in dendritic arborization and spine morphology of developing pyramidal neurons. J. Neurosci. 2016;36:2723–2742. doi: 10.1523/JNEUROSCI.2321-15.2016. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 40. Moffat J.J., Jung E.M., Ka M., Smith A.J., Jeon B.T., Santen G.W., Kim W.Y. The role of ARID1B, a BAF chromatin remodeling complex subunit, in neural development and behavior. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;89:30–38. doi: 10.1016/j.pnpbp.2018.08.021. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 41. Moffat J.J., Jung E.M., Ka M., Jeon B.T., Lee H., Kim W.Y. Differential roles of ARID1B in excitatory and inhibitory neural progenitors in the developing cortex. Sci. Rep. 2021;11:3856. doi: 10.1038/s41598-021-82974-y. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 42. Kim H., Kim D., Cho Y., Kim K., Roh J.D., Kim Y., Yang E., Kim S.S., Ahn S., Kim H., et al. Early postnatal serotonin modulation prevents adult-stage deficits in Arid1b deficient mice through synaptic transcriptional reprogramming. Nat. Commun. 2022;13:5051. doi: 10.1038/s41467-022-32748-5. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 43. Moffat G., Zhukovsky P., Coughan G., Voineskos A.N. Unravelling the relationship between amyloid accumulation and brain network function in normal aging and very mild cognitive decline: A longitudinal analysis. Brain Commun. 2022;4:fcac282. doi: 10.1093/braincomms/fcac282. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 44. Nelson A.D., Bender K.J. Dendritic integration dysfunction in neurodevelopment disorders. Dev. Neurosci. 2021;43:201–221. doi: 10.1159/000516657. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 45. Lee K., Vyas Y., Garner C.C., Montgomery J.M. Autism associated Shank3 mutations alter mGluR expression and mGluR-dependent NMDA receptor-dependent long-term depression. Synapse. 2019;73:222097. doi: 10.1002/syn.22097. [ DOI ] [ PubMed ] [ Google Scholar ]
- 46. Tian C., Psukus J.D., Fingeton E., Roche K.W., Herring B.E. Autism spectrum disorder intellectual disability-associated mutations in trio disrupt neuroligin 1-mediated synaptogenesis. J. Neurosci. 2021;41:7768–7778. doi: 10.1523/JNEUROSCI.3148-20.2021. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 47. Hsieh M.Y., Tuan L.H., Chang H.C., Wang Y.C., Chen C.H., Shy H.C., Lee L.J., Shur-Fen Gau S. Altered synaptic protein expression, aberrant spine morphology, and impaired spatial memory in Digap2 mutant mice, a genetic model of autism spectrum disorder. Cereb. Cortex. 2022;28:bhac379. doi: 10.1093/cercor/bhac379. [ DOI ] [ PubMed ] [ Google Scholar ]
- 48. Xie Y., Wang H., Hu B., Zhang X., Liu A., Cai C., Li S., Chen C., Wang Z., Yin Z., et al. Dendritic spine in autism genetics: Whole-exome sequencing identifying de novo variants of CTTNBP2 in a quad family affected by autism spectrum disorder. Children. 2022;10:80. doi: 10.3390/children10010080. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 49. Urrutia-Ruiz C., Rombach D., Cursano S., Gerlach-Arbeiter S., Schoen M., Bockmann J., Demestre M., Boeckers T.M. Deletion of the autism-associated protein SHANK3 abolishes structural synaptic plasticity after brain trauma. Int. J. Mol. Sci. 2022;23:6081. doi: 10.3390/ijms23116081. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 50. Bucher M., Niebling S., Han Y., Molodenskiy D., Hassani-Nia F., Kreinkamp H.J., Svergun D., Kim E., Kostiukova A.S., Kreutz M.R. Autism associated SHANK3 missense point mutation impact conformational fluctuations and protein turnover at synapses. eLife. 2021;10:e66165. doi: 10.7554/eLife.66165. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 51. Nishiyama J. Plasticity of dendritic spines: Molecular function and dysfunction in neurodevelopmental disorders. Psychiatry Clin. Neurosci. 2019;73:541–550. doi: 10.1111/pcn.12899. [ DOI ] [ PubMed ] [ Google Scholar ]
- 52. Lamanna J., Isotti F., Ferro M., Spedini S., Racchetti R., Musazzi L., Malgaroli A. Occlusion of dopamine-dependent synaptic plasticity in the prefrontal cortex mediates the expression of depressive-like behavior and is medulaed by ketamine. Sci. Rep. 2022;12:11055. doi: 10.1038/s41598-022-14694-w. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 53. Kumar Tripathi M., Kumar Oiha S., Kartawy M., Hamoundi W., Choudhary A., Stern S., Aran A., Amal H. The NO answer for autism spectrum disorder. Adv. Sci. (Weinh) 2023;10:e2205783. doi: 10.1002/advs.202205783. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 54. Porokhovnik L.N., Pisarev V.M., Chumachenko A.G., Chudkova J.M., Ershova E.S., Veiko N.N., Gorbachevskaya N.L., Mamokhina U.A., Sorokin A.B., Basova A.Y. Association of NEF2L2 Rs35652124 polymorphism with Nrf2 induction and genotoxic stress biomarkers in autism. Gene. 2023;14:718. doi: 10.3390/genes14030718. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 55. Memis I., Mittal R., Furar E., White I., Eshraghi A.A. Altered blood brain barrier permeability and oxidative stress in Cntnap2 knockout rat model. J. Clin. Med. 2022;11:2725. doi: 10.3390/jcm11102725. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 56. Petroni V., Subashi E., Premoli M., Wohr M., Crusio W.E., Memaire V., Pietropaolo S. Autistic-like behavioral effects of prenatal stress in juvenile Fmr1 mice: The relevance of sex differences and gene environment interactions. Sci. Rep. 2022;12:7269. doi: 10.1038/s41598-022-11083-1. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 57. Kilpatrick S., Irwin C., Singh K.K. Human pluripotent stem cells (hPSC) and organoid models of autism: Opportunities and limitations. Transl. Psychiatry. 2023;13:217. doi: 10.1038/s41398-023-02510-6. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 58. Vakilzadeh G., Falcone C., Dufour B., Hong T., Noctor S.C., Martinez-Cerdeno V. Decreased number and increased activation state of astrocytes in gray and white matter of the prefrontal cortex in autism. Cereb. Cortex. 2022;32:4902–4912. doi: 10.1093/cercor/bhab523. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 59. Allen M., Huang B.S., Notaras M.J., Lodhi A., Barrio-Alonso E., Lituma P.J., Wolujewicz P., Wiztum J., Longo F., Chen M., et al. Astrocytes derived from ASD individuals alter behavior and destabilize neuronal activity through aberrant Ca2+ signaling. Mol. Psychiatry. 2022;27:2470–2484. doi: 10.1038/s41380-022-01486-x. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 60. Vakilzadeh G., Martinez-Cerdeno V. Pathology and astrocytes in autism. Neuropsychiatr. Dis. Treat. 2023;19:841–850. doi: 10.2147/NDT.S390053. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 61. Guneykaya D., Uguru B., Logiacco P., Popp O., Feiks M.A., Meyer N., Wendt S., Semter M., Cherif F., Gauthier C., et al. Sex-specific microglia state in neuroligin-4 knock-out mouse model of autism spectrum disorder. Brain Behav. Immunol. 2023;111:61–75. doi: 10.1016/j.bbi.2023.03.023. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 62. Luo Y., Lv K., Du Z., Zhang D., Chen M., Luo J., Wang L., Liu T., Gong H., Fan X. Monocycline improves autism-related behaviors by modulating microglia polarization in mouse model autism. Int. Immunopharmacol. 2023;122:110594. doi: 10.1016/j.intimp.2023.110594. [ DOI ] [ PubMed ] [ Google Scholar ]
- 63. Xiong Y., Chen J., Li Y. Microglia and astrocytes underlie neuroinflammation and synaptic susceptibility in autism spectrum disorder. Front. Neurosci. 2023;17:1125428. doi: 10.3389/fnins.2023.1125428. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 64. Frasch M.G., Yoon B.J., Helbing D.L., Snir G., Antonelli M.C., Bauer R. Autism spectrum disorder: A neuro-immunometabolic hypothesis of the developmental origins. Biology. 2023;12:914. doi: 10.3390/biology12070914. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 65. Usui N., Kobayashi H., Shimida S. Neuroinflammation and oxidative stress in the pathogenesis of autism spectrum disorder. Int. J. Mol. Sci. 2023;24:5487. doi: 10.3390/ijms24065487. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 66. Costa A.N., Ferguson B.J., Hawkins E., Coman A., Schauer J., Ramirez-Cells A., Hecht P.M., Bruce D., Tilley M., Zelebizadeh Z., et al. Relationship between maternal antibodies in fetal brain and pre-natal stress exposure in autism spectrum disorder. Metabolites. 2023;13:663. doi: 10.3390/metabo13050663. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 67. Woo T., King C., Ahmed N.I., Cordes M., Nistala S., Will M.J., Bloomer C., Kibiryev N., Rivera M.R., Telebizadeh Z., et al. microRNA as a maternal marker for the prenatal stress-associated ASD, evidence from a murine model. J. Pers. Med. 2023;13:1412. doi: 10.3390/jpm13091412. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 68. Harrison Elder J., Maun Kreider C., Brasher S.N., Ansell M. Clinical impact of early diagnosis of autism on the prognosis and parent-child relationships. Psychol. Res. Behav. Manag. 2017;10:283–292. doi: 10.2147/PRBM.S117499. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 69. Klin A. Translating advances in developmental social neuroscience into greater access to early diagnosis in autism spectrum disorder. Medicine. 2023;83((Suppl. 2)):32–36. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 70. Costantino J.N., Abbacchi A.M., May B.K., Klaiman C., Zhang Y., Lowe J.K., Marrus N., Klin A., Geschwin D.H. Prospects for leveling the playing field for black children with autism. J. Am. Acad. Child Adolesc. Psychiatry. 2023;62:949–952. doi: 10.1016/j.jaac.2023.05.005. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 71. Jones W., Klaiman C., Richardson S., Lambha M., Reid M., Hamnes T., Beacham C., Lewis P., Paredes J., Edwards L., et al. Development and replication of objective measurements of social visual engagement to aid in early diagnosis and assessment of autism. JAMA Netw. Open. 2023;6:e2330145. doi: 10.1001/jamanetworkopen.2023.30145. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 72. Sobieski M., Sobieska A., Sekulowicz M., Bujnowska-Fedak M.M. Tools for early screening of autism spectrum disorders in primary health care- a scoping review. BMC Prim. Care. 2022;23:46. doi: 10.1186/s12875-022-01645-7. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 73. Klaiman C., White S., Richardson S., McQueen E., Walum H., Aoki C., Smith C., Minjarez M., Bernier R., Pedapati E., et al. Expert clinician certainty in diagnosing autism spectrum disorder in 16-30 months old: A multi-site trial secondary analysis. J. Autism Dev. Disord. 2022;17:1–16. doi: 10.1007/s10803-022-05812-8. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 74. Roy M., Strate P. Autism spectrum disorders in adulthood-symptoms diagnosis. Dtsch. Arztebl. Int. 2023;120:87–93. doi: 10.3238/arztebl.m2022.0379. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 75. Angel L., Alley S.H., Delaney K.R., Mohr L. Presentation of depressive symptoms in autism spectrum disorders. West. J. Nurs. Res. 2023;45:854–861. doi: 10.1177/01939459231190269. [ DOI ] [ PubMed ] [ Google Scholar ]
- 76. Moffat J.J., Smith A.L., Jung A.L., Ka M., Kim W.Y. Neurobiology of ARID1B haploinsufficiency related to neurodevelopmental and psychiatric disorders. Mol. Psychiatry. 2022;27:476–489. doi: 10.1038/s41380-021-01060-x. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 77. Grzadzinski R., Dick C., Lord C., Bishop S. Parent-reported and clinician-observed autism spectrum disorder (ASD) symptoms in children with attention deficit/hyperactivity disorder (ADHD): Implications for practice. Mol. Autism. 2016;7:7. doi: 10.1186/s13229-016-0072-1. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 78. Wu M., Jubran E., Kumar D., Assad N.D., Nguyen H. Variations in anxiety and related in psychiatric comorbidity levels among youth with individual diagnoses of autism spectrum disorder or attention deficit hyperactivity disorder and those with both diagnoses. Cureus. 2023;15:e41759. doi: 10.7759/cureus.41759. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 79. Mannion A., Leader G. Relationship between child sleep problems in autism spectrum disorder and parent mental health and well-being. Sleep Med. 2023;109:4–10. doi: 10.1016/j.sleep.2023.05.009. [ DOI ] [ PubMed ] [ Google Scholar ]
- 80. Mai A.S., Yau C.E., Tzeng F.S., Foo Q.X.J., Wang D.Q., Tan E.-K. Linking autism spectrum disorders and parkinsonism: Clinical and genetic association. Ann. Clin. Transl. Neurol. 2023;10:484–496. doi: 10.1002/acn3.51736. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 81. Frederiksen S.D., Wicki-Stordeur L.E., Swayne L.A. Overlap in synaptic neurological conditions susceptibility pathways and the neuronal pannexin 1 interactome revealed by bioinformatics analyses. Channels. 2023;17:2253102. doi: 10.1080/19336950.2023.2253102. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 82. Das M., Zomorrodi R., Mirialli M., Lirkovski M., Blumbrg D.M., Rajj T.K., Desarkar P. Machine learning approaches for electroencephalography and magnetoencephaligraphy analyses in autism spectrum disorder. Progr. Neuropsychopharmacol. Biol. Psychiatry. 2023;123:110705. doi: 10.1016/j.pnpbp.2022.110705. [ DOI ] [ PubMed ] [ Google Scholar ]
- 83. Lamy M., Erickson C.A. Pharmacological management of behavioral disturbances in children and adolescents with autism spectrum disorders. Curr. Probl. Pediatr. Adolesc. Health Care. 2018;48:250–264. doi: 10.1016/j.cppeds.2018.08.015. [ DOI ] [ PubMed ] [ Google Scholar ]
- 84. Aran A., Rand D.C. Cannabinoid treatment for the symptoms of autism spectrum disorder. Expert Opin. Emerg. Drugs. 2024:1–15. doi: 10.1080/14728214.2024.2306290. [ DOI ] [ PubMed ] [ Google Scholar ]
- 85. Rahim F., Toguzbaeva K., Quasim N.H., Dzhusupov K.O., Zuhmagaliuly A., Koozhamkul R. Probiotics, prebiotics, and synbiotics for patients with autism spectrum disorder: A meta analysis and umbrella review. Front Nutr. 2023;10:1294089. doi: 10.3389/fnut.2023.1294089. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 86. Wichers R.H., wan der Wouw L.C., Brouwer M.E., Lok A., Bockting C.L.H. Psychotherapy for co-occurring symptoms of depression, anxiety and obsessive-compulsive disorder in children and adults with autism spectrum disorder: A systematic review and meta-analysis. Psychol. Med. 2023;53:17–33. doi: 10.1017/S0033291722003415. [ DOI ] [ PubMed ] [ Google Scholar ]
- 87. Yeung P.P., Johnson K.A., Riesenberg R., Orejudos A., Riccobene T., Kalluri H.V., Malik P.R., Varughese S. Carioprazine in pediatric patients with autism spectrum disorder: Results of a pharmacokinetic, safety and tolerability study. J. Child Adolesc. Psychopharmacol. 2023;33:232–242. doi: 10.1089/cap.2022.0097. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 88. Valdovinos M.G., Epperson C., Johnson C. A review of the use of psychotropic medication address challenging behaviour in neurodevelopmental disorders. Int. Rev. Neurobiol. 2023;173:43–65. doi: 10.1016/bs.irn.2023.08.010. [ DOI ] [ PubMed ] [ Google Scholar ]
- 89. Arteaga-Henriquez G., Gisbeert L., Ramos Quiroga J.A. Immunoregulatory and/or anti-inflammatory agents for the management of core and associated symptoms in individuals with autism spectrum disorder: A narrative review of randomized, placebo-controlled trials. CNS Drugs. 2023;37:215–229. doi: 10.1007/s40263-023-00993-x. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 90. Caldani S., Humeau E., Deorme R., Bucci M.P. Inhibition functions can be improved in children with autism spectrum disorders: An eye-tracking study. Int. J. Dev. Neurosci. 2023;83:431–441. doi: 10.1002/jdn.10276. [ DOI ] [ PubMed ] [ Google Scholar ]
- 91. Patel M., Atluri M.L., Gonzalez M.A., Sakhamuri N., Athiyaman S., Randhi B., Gutlpalli S.D., Pu J., Faldi M.F., Khan S., et al. Systematic review of mixed studies exploring the effects of probiotics on gut-microbiome to modulate therapy in children with autism spectrum disorder. Cureus. 2022;14:e3213. doi: 10.7759/cureus.32313. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 92. Yu Z., Zhang P., Tao C., Lu L., Tang C. Efficacy of non-pharmacological interventions targeting social functions in children and adults with autism spectrum disorder: A systematic review and meta-analysis. PLoS ONE. 2023;18:e0291720. doi: 10.1371/journal.pone.0291720. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 93. Hellings J. Pharmacotherapy in autism spectrum disorder, including promising drugs warranting trials. World J. Psychiatry. 2023;13:262–277. doi: 10.5498/wjp.v13.i6.262. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 94. van der Plas E., Mason D., Happé F. Decision making in autism: A narrative review. Autism. 2023;27:1532–1546. doi: 10.1177/13623613221148010. [ DOI ] [ PubMed ] [ Google Scholar ]
- 95. Adaralegbe N.J., Okobi O.E., Omar Z.T.O., Segun E., Evbayekha E.O., Abolurin A., Egberuare E.O., Ezegbe H.C., Adegbosin A., Adedeji A.G., et al. Impact of adverse childhood experience on resilience and school success in individuals with autism spectrum disorder and attention-deficit hyperactivity disorder. Cureus. 2022;14:e31907. doi: 10.7759/cureus.31907. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 96. Tayanloo-Beik A., Hamidpour S.K., Abedi M., Shojaei H., Tavirani M.R., Namzi N., Larijani B., Arjmand D. Zebrafish modeling of autism spectrum disorders, current status and future prospective. Front. Psychiatry. 2022;13:911770. doi: 10.3389/fpsyt.2022.911770. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 97. Etkin R.G., Juel E.K., Lebwitz E.R., Silverman W. Does cognitive-behavioral therapy for youth anxiety disorders improve social functioning and peer relationships? Clin. Child Fam. Psychol. Rev. 2023;26:1052–1076. doi: 10.1007/s10567-023-00454-3. [ DOI ] [ PubMed ] [ Google Scholar ]
- 98. Saha S.K., Joshi A., Singh R., Dubey K. Review of industrially recognized polymers and manufacturing processes for amorphous solid dispersion based formulations. Pharm. Dev. Technol. 2023;28:678–696. doi: 10.1080/10837450.2023.2233595. [ DOI ] [ PubMed ] [ Google Scholar ]
- View on publisher site
- PDF (10.1 MB)
- Collections
Similar articles
Cited by other articles, links to ncbi databases.
- Download .nbib .nbib
- Format: AMA APA MLA NLM
Add to Collections

IMAGES
COMMENTS
May 23, 2024 · Geschwind's study on autism, one of nine published in the May 24 issue of Science, builds on decades of his group's research profiling the genes that increase the susceptibility to autism spectrum ...
Jul 30, 2023 · Most genetic studies of autism have focused on families with one child affected by the neurodevelopmental disorder, sometimes excluding families with multiple affected children. As a result, few studies have examined the role of rare inherited variation or its interaction with the combined effect of multiple common genetic variations that ...
5 days ago · 2024 brought important progress in autism research, with discoveries that deepen our understanding of autism and open new possibilities for improving care. Advances in genetic research, technology and diagnostic tools are paving the way for better outcomes for autistic people and their families.
Aug 10, 2023 · Modeling idiopathic autism in forebrain organoids reveals an imbalance of excitatory cortical neuron subtypes during early neurogenesis. Nature Neuroscience , 2023; DOI: 10.1038/s41593-023-01399-0 ...
Decades of research have since revealed that autism is a highly heterogeneous and extremely complex genetic condition. Even though great progress had been made in identifying hundreds of risk genes, very little is known about the different types of modifiers that may exacerbate or ameliorate disease severity.
4 days ago · New research published in The American Journal of Human Genetics has identified a previously unknown genetic link to autism spectrum disorder (ASD). The study found that variants in the DDX53 gene ...
5 days ago · Background Autism spectrum disorder (ASD) is a partially heritable neurodevelopmental trait, and people with ASD may also have other co-occurring trait such as ADHD, anxiety disorders, depression, mental health issues, learning difficulty, physical health traits and communication challenges. The concomitant development of ASD and other neurological traits is assumed to result from a complex ...
4 days ago · The high prevalence and high rank for non-fatal health burden of autism spectrum disorder in people younger than 20 years underscore the importance of early detection and support to autistic young people and their caregivers globally. Work to improve the precision and global representation of our findings is required, starting with better global coverage of epidemiological data so that ...
The disease was discovered 75 years ago, and the present review, based on critical evaluations of the recognized ASD studies from the beginning of 1990, has been further developed by the comparative analyses of the research and clinical reports, which have grown progressively in recent years up to late 2023.
Sep 28, 2015 · In the largest, most comprehensive genomic analysis of autism spectrum disorder (ASD) conducted to date, an international research team led by UCSF Psychiatry scientists has identified 65 genes that play a role in the disorder, 28 of which are reported with “very high confidence,” meaning that there is 99 percent certainty that these genes contribute to the risk of developing ASD.